CellBench Data
Kevin Wang
12 Apr 2024
Source:vignettes/case_study/CellBench_Data.Rmd
CellBench_Data.RmdIntroduction
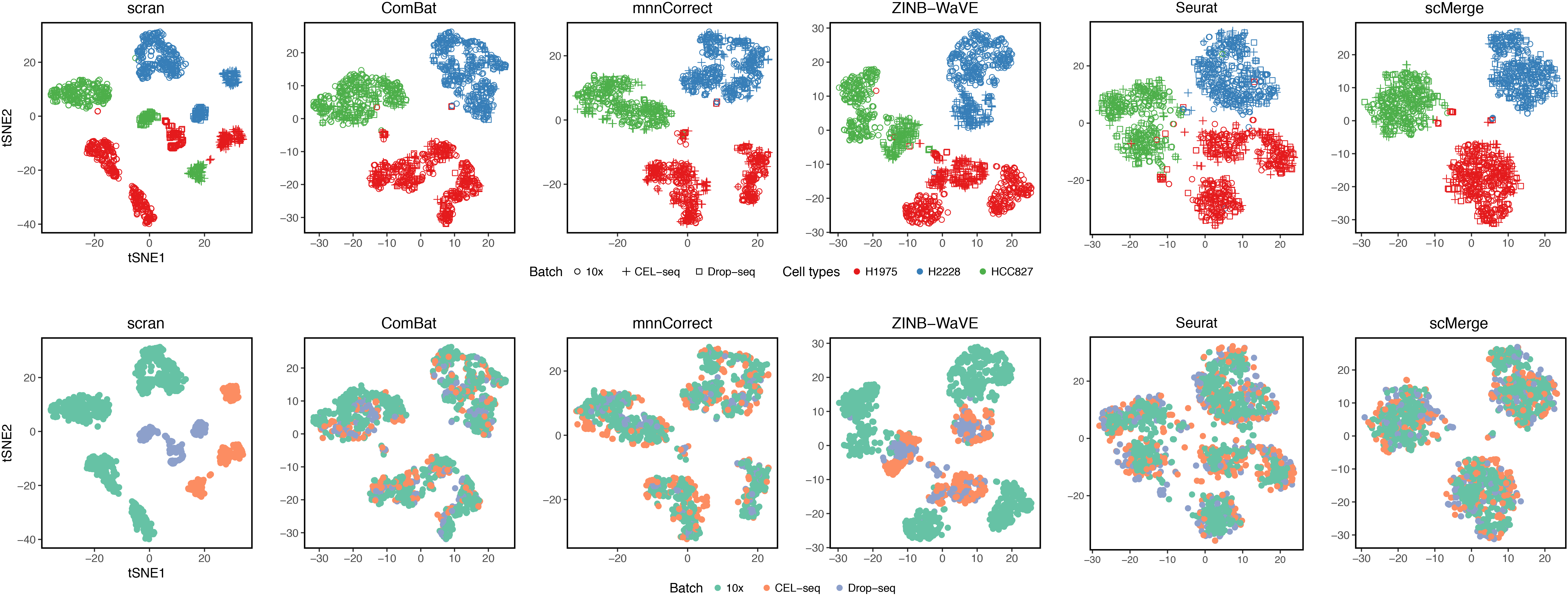
This is a benchmarking single-cell data comprising of 3 different human lung adenocarcinoma cell lines. This experiment was designed induce different batch effects into the data.
Integration challenge
- Prior to integration, there is a strong separation effect by multiple batch effects.
- One of the batch effects is the different protocols with significant depth differences.
- The experimental design poses extra difficulties as cell lines were cultured separatedly and the same batch was processed in 3 different ways.
Data description
- Data source:
| Type of merge | Name | ID | Author | DOI or URL | Protocol | Organism | Tissue | # of cell types | # of cells | # of batches | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Across platforms with significant depth difference | CellBench | CellBench | https://github.com/LuyiTian/CellBench_data | Cel-seq2, Drop-seq, 10x Chromium | Human | Adenocarcinoma cell lines | 3 | 1401 | 3 per cell types |
- Relation to the
scMergearticle: Supplementary Figure 12.
Integrated scMerge data
Data availability: CellBench Data (in RData format)
-
scMergeparameters for integration:- Unsupervised scMerge
- kmeans K = (3,3,3)
- Negative controls are human scSEG