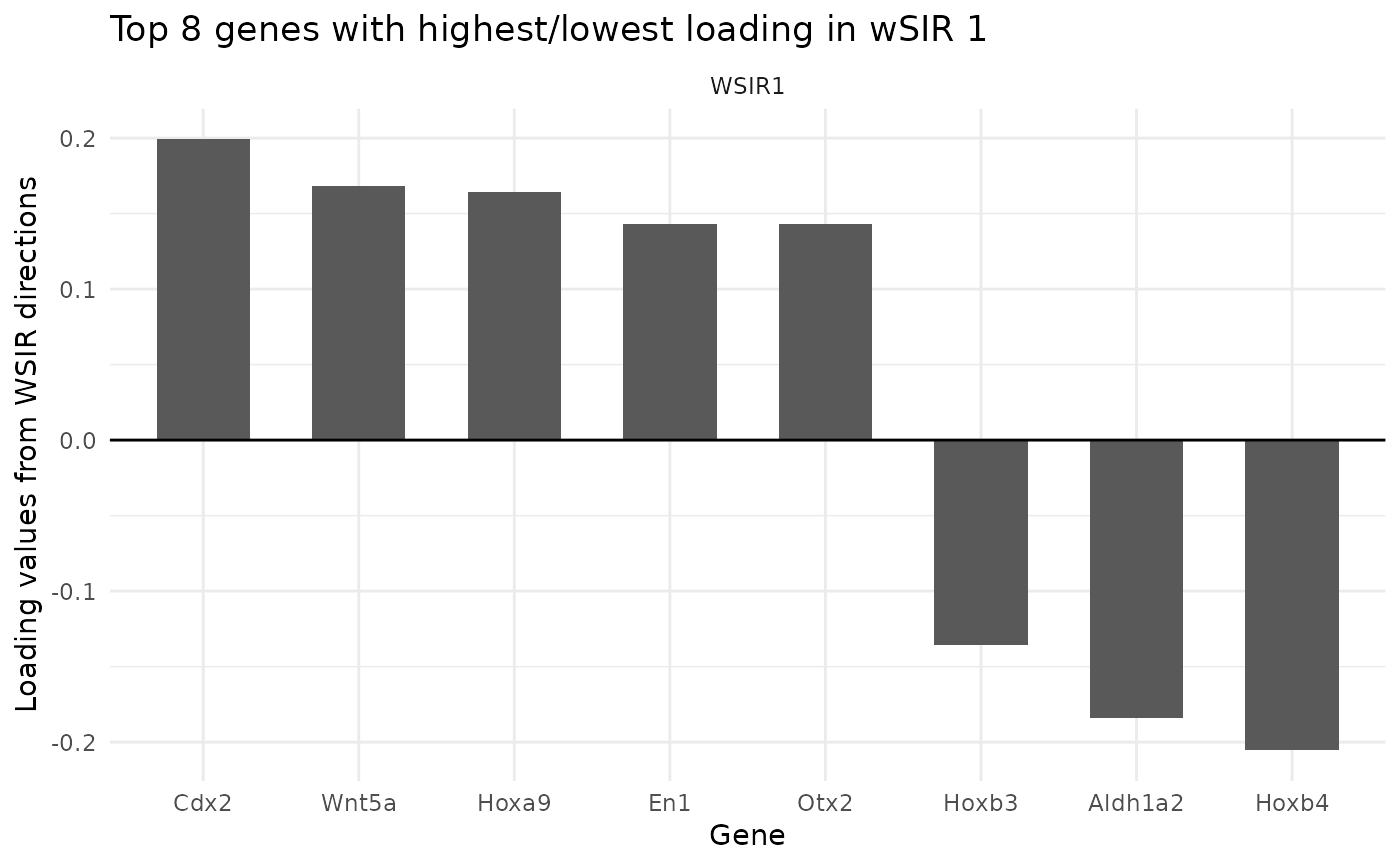
A function to find and visualise the genes with the highest
(in absolute value) loading in WSIR1.
These genes contribute the most to the first low-dimensional direction.
Usage
findTopGenes(WSIR, highest = 10, dirs = 1)
Arguments
- WSIR
wsir object as output of wSIR function. To analyse a different
DR method, ensure the
slot named 'directions' contains the loadings as a matrix with the gene
names as the rownames.
- highest
integer for how many of the top genes you would like to see.
Recommend no more
than 20 for ease of visualisation. Default is 10.
- dirs
integer or vector for which direction / directions you want to
show/find the top genes from.
Value
List containing two slots. First is named "plot" and shows a
barplot with the top genes
and their corresponding loadings. Second is named "genes" and is a vector
of the genes with highest
(in absolute value) loading in low-dimensional direction 1. Length is
parameter highest.
Examples
data(MouseData)
wsir_obj = wSIR(X = sample1_exprs,
coords = sample1_coords,
optim_params = FALSE,
alpha = 4,
slices = 6) # create wsir object
top_genes_obj = findTopGenes(WSIR = wsir_obj, highest = 8)
top_genes_plot = top_genes_obj$plot # select plot
top_genes_plot # print plot