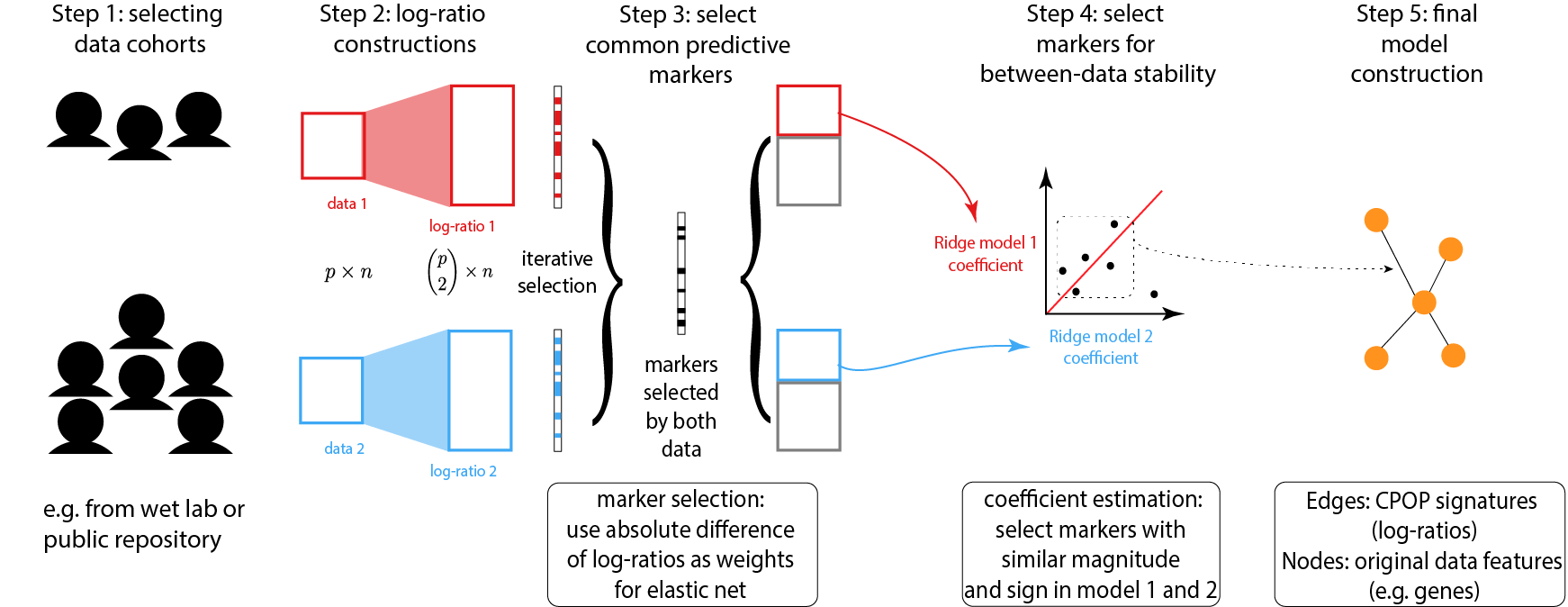
CPOP - Cross-Platform Omics Prediction
Due to data scale differences between multiple omics data, a model constructed from a training data tends to have poor prediction power on a validation data. While the usual bioinformatics approach is to re-normalise both the training and the validation data, this step may not be possible due to ethics constrains. CPOP avoids re-normalisation of additional data through the use of log-ratio features and thus also enable prediction for single omics samples.
The novelty of the CPOP procedure lies in its ability to construct a transferable model across gene expression platforms and for prospective experiments. Such a transferable model can be trained to make predictions on independent validation data with an accuracy that is similar to a re-substituted model. The CPOP procedure also has the flexibility to be adapted to suit the most common clinical response variables, including linear response, binomial and Cox PH models.
References
Cross-Platform Omics Prediction procedure: a statistical machine learning framework for wider implementation of precision medicine Kevin Y.X. Wang, Gulietta M. Pupo, Varsha Tembe, Ellis Patrick, Dario Strbenac, Sarah-Jane Schramm, John F. Thompson, Richard A. Scolyer, Samuel Mueller, Garth Tarr, Graham J. Mann, Jean Y.H. Yang bioRxiv 2020.12.09.415927; doi: https://doi.org/10.1101/2020.12.09.415927