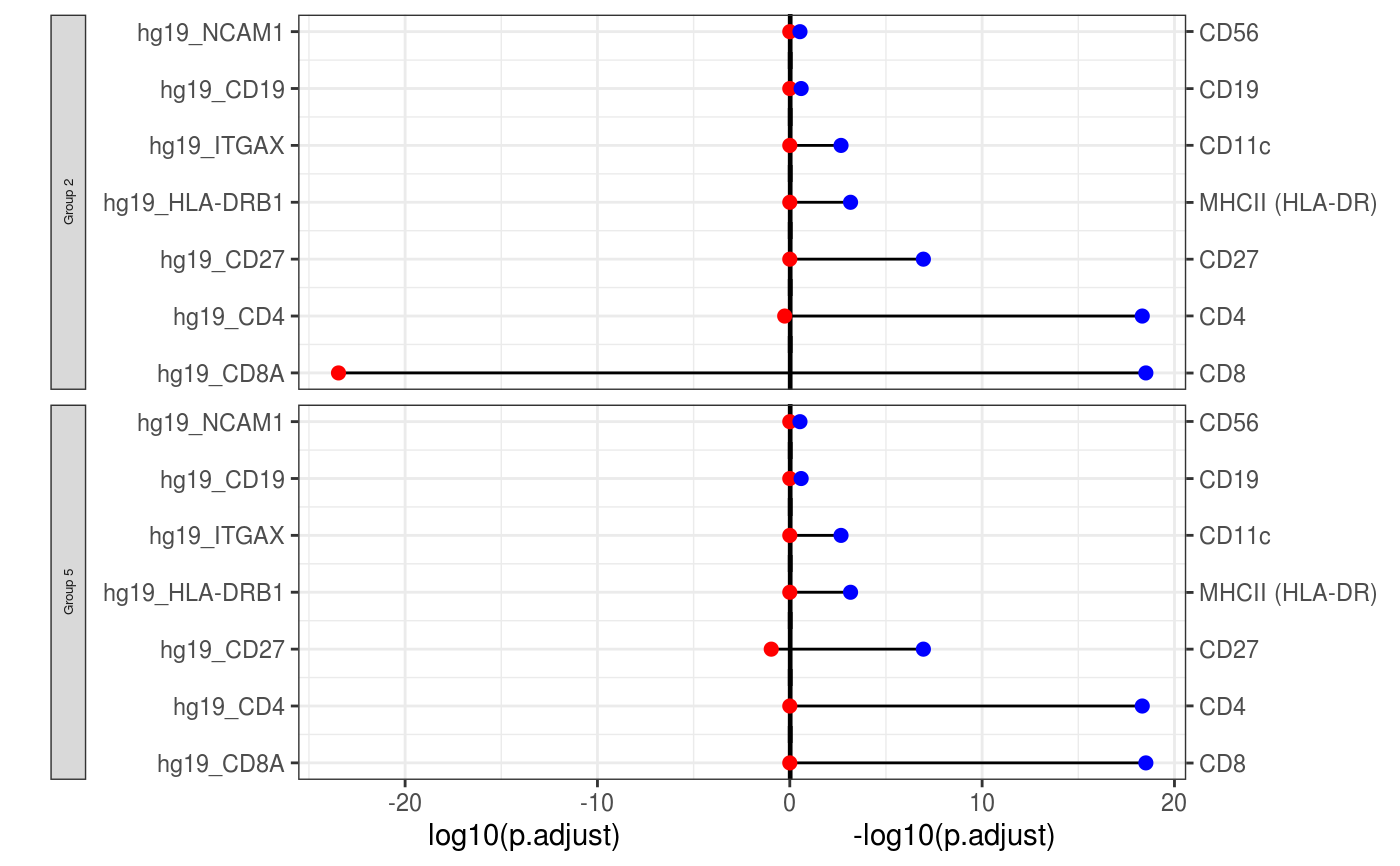
A function to visualise the pairwise comparison of pvalue in different data modality.
DEcomparisonPlot(de_list, feature_list)
Arguments
| de_list | A list including two lists results from `DE genes ()`. |
|---|---|
| feature_list | A list including two lists features indicating the selected subset of features will be visualised |
Value
A ggplot2 to visualise the comparison plot of DE.
Examples
library(S4Vectors) data(sce_control_subset) sce_control_subset <- DEgenes(sce_control_subset, group = sce_control_subset$SNF_W_louvain, return_all = TRUE, exprs_pct = 0.5) sce_control_subset <- selectDEgenes(sce_control_subset) sce_control_subset <- DEgenes(sce_control_subset, altExp_name = "ADT", group = sce_control_subset$SNF_W_louvain, return_all = TRUE, exprs_pct = 0.5) sce_control_subset <- selectDEgenes(sce_control_subset, altExp_name = "ADT") rna_list <- c("hg19_CD4", "hg19_CD8A", "hg19_HLA-DRB1", "hg19_ITGAX", "hg19_NCAM1", "hg19_CD27", "hg19_CD19") adt_list <- c("CD4", "CD8", "MHCII (HLA-DR)", "CD11c", "CD56", "CD27", "CD19") rna_DEgenes_all <- S4Vectors::metadata(sce_control_subset)[["DE_res_RNA"]] adt_DEgenes_all <- S4Vectors::metadata(sce_control_subset)[["DE_res_ADT"]] feature_list <- list(RNA = rna_list, ADT = adt_list) de_list <- list(RNA = rna_DEgenes_all, ADT = adt_DEgenes_all) DEcomparisonPlot(de_list = de_list, feature_list = feature_list)#> Warning: Use of `df$group` is discouraged. Use `group` instead.#> Warning: Use of `df$group` is discouraged. Use `group` instead.#> Warning: Use of `df$group` is discouraged. Use `group` instead.#> Warning: Use of `df$group` is discouraged. Use `group` instead.#> Warning: Use of `df$group` is discouraged. Use `group` instead.