A function to visualise the features distribtuion
visualiseExprs( sce, plot = c("boxplot", "violin", "jitter", "density", "pairwise"), altExp_name = c("none"), exprs_value = "logcounts", group_by = NULL, facet_by = NULL, feature_subset = NULL, cell_subset = NULL, n = NULL, threshold = NULL )
Arguments
| sce | A singlecellexperiment object |
|---|---|
| plot | Type of plot, includes boxplot, violin, jitter, density, and pairwise. By default is boxplot |
| altExp_name | A character indicates which expression matrix is used. by default is none (i.e. RNA). |
| exprs_value | A character indicates which expression value in assayNames is used. |
| group_by | A character indicates how is the expression will be group in the plots (stored in colData). |
| facet_by | A character indicates how is the expression will be lay out panels in a grid in the plots (stored in colData). |
| feature_subset | A vector of characters indicates the subset of features that are used for visualisation |
| cell_subset | A vector of characters indicates the subset of cells that are used for visualisation |
| n | A numeric indicates the top expressed features to show. |
| threshold | Thresholds of high expresion for features (only is used for pairwise plot). |
Value
A ggplot to visualise te features distribution
Examples
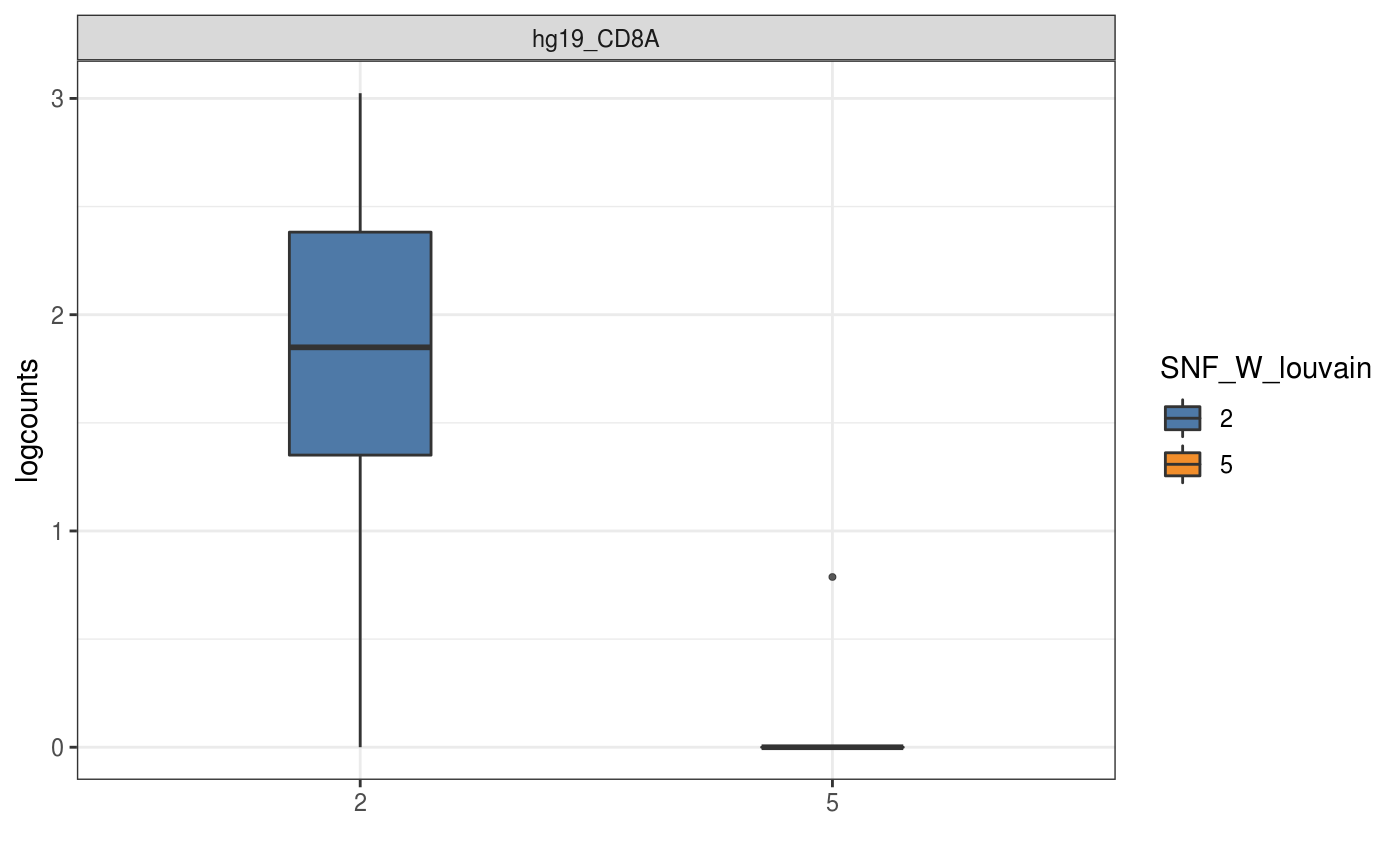
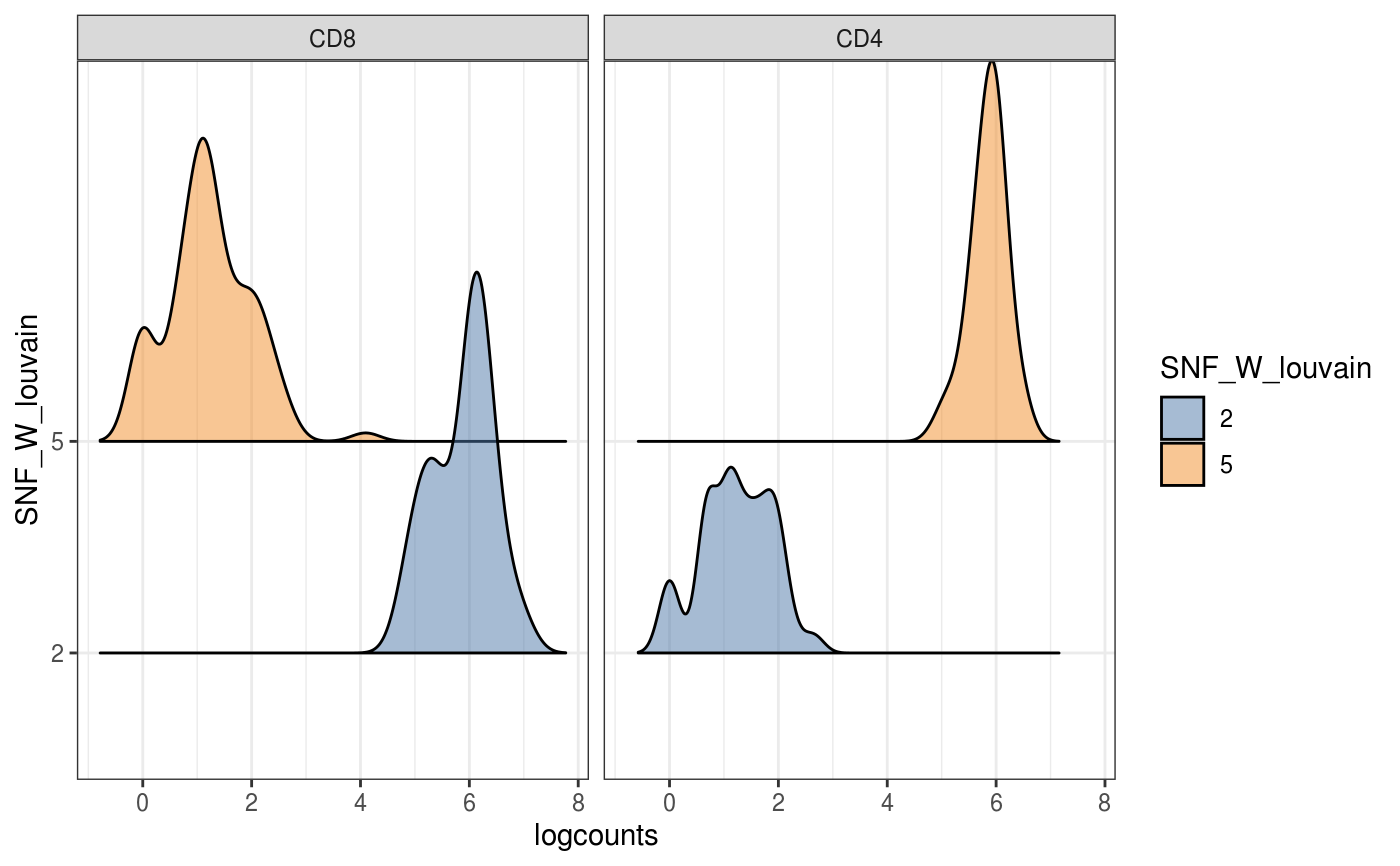
data(sce_control_subset) visualiseExprs(sce_control_subset, plot = "boxplot", group_by = "SNF_W_louvain", feature_subset = c("hg19_CD8A"))visualiseExprs(sce_control_subset, plot = "density", altExp_name = "ADT", group_by = "SNF_W_louvain", feature_subset = c("CD8", "CD4"))#>#>