Cross-validation to evaluate classification performance.
crossValidate.RdThis function has been designed to facilitate the comparison of classification
methods using cross-validation, particularly when there are multiple assays per biological unit.
A selection of typical comparisons are implemented. The train function
is a convenience method for training on one data set and likewise predict for predicting on an
independent validation data set.
Usage
# S4 method for class 'DataFrame'
crossValidate(
measurements,
outcome,
nFeatures = 20,
selectionMethod = "auto",
classifier = "auto",
multiViewMethod = "none",
assayCombinations = "all",
nFolds = 5,
nRepeats = 20,
nCores = 1,
characteristicsLabel = NULL,
extraParams = NULL,
verbose = 0
)
# S4 method for class 'MultiAssayExperimentOrList'
crossValidate(
measurements,
outcome,
nFeatures = 20,
selectionMethod = "auto",
classifier = "auto",
multiViewMethod = "none",
assayCombinations = "all",
nFolds = 5,
nRepeats = 20,
nCores = 1,
characteristicsLabel = NULL,
extraParams = NULL,
verbose = 0
)
# S4 method for class 'data.frame'
crossValidate(
measurements,
outcome,
nFeatures = 20,
selectionMethod = "auto",
classifier = "auto",
multiViewMethod = "none",
assayCombinations = "all",
nFolds = 5,
nRepeats = 20,
nCores = 1,
characteristicsLabel = NULL,
extraParams = NULL,
verbose = 0
)
# S4 method for class 'matrix'
crossValidate(
measurements,
outcome,
nFeatures = 20,
selectionMethod = "auto",
classifier = "auto",
multiViewMethod = "none",
assayCombinations = "all",
nFolds = 5,
nRepeats = 20,
nCores = 1,
characteristicsLabel = NULL,
extraParams = NULL,
verbose = 0
)
# S3 method for class 'matrix'
train(x, outcomeTrain, ...)
# S3 method for class 'data.frame'
train(x, outcomeTrain, ...)
# S3 method for class 'DataFrame'
train(
x,
outcomeTrain,
selectionMethod = "auto",
nFeatures = 20,
classifier = "auto",
multiViewMethod = "none",
assayIDs = "all",
extraParams = NULL,
verbose = 0,
...
)
# S3 method for class 'list'
train(x, outcomeTrain, ...)
# S3 method for class 'MultiAssayExperiment'
train(x, outcome, ...)
# S3 method for class 'trainedByClassifyR'
predict(object, newData, outcome, ...)Arguments
- measurements
Either a
DataFrame,data.frame,matrix,MultiAssayExperimentor a list of the basic tabular objects containing the data.- outcome
A vector of class labels of class
factorof the same length as the number of samples inmeasurementsor a character vector of length 1 containing the column name inmeasurementsif it is aDataFrame. Or aSurvobject or a character vector of length 2 or 3 specifying the time and event columns inmeasurementsfor survival outcome. Ifmeasurementsis aMultiAssayExperiment, the column name(s) incolData(measurements)representing the outcome. If column names of survival information, time must be in first column and event status in the second.- ...
For
trainandpredictfunctions, parameters not used by the non-DataFrame signature functions but passed into the DataFrame signature function.- nFeatures
The number of features to be used for classification. If a named vector with the same names of multiple assays, a different number of features will be used for each assay. Set to
"all"if all features should be used. To tune it, specify a vector orlistof named vectors to"tuneParams"list of"select"element list ofextraParamslist.- selectionMethod
Default:
"auto". A character vector of feature selection methods to compare. If a named character vector with names corresponding to different assays, and performing multiview classification, the respective selection methods will be used on each assay. If"auto", t-test (two categories) / F-test (three or more categories) ranking and topnFeaturesoptimisation is done. Otherwise, the ranking method is per-feature Cox proportional hazards p-value."none"is also a valid value, meaning that no feature selection prior to model building will be performed (but implicit selection might still happen with the classifier).- classifier
Default:
"auto". A character vector of classification methods to compare. If a named character vector with names corresponding to different assays, and performing multiview classification, the respective classification methods will be used on each assay. If"auto", then a random forest is used for a classification task or Cox proportional hazards model for a survival task.- multiViewMethod
Default:
"none". A character vector specifying the multiview method or data integration approach to use. Seeavailable("multiViewMethod") for possibilities.- assayCombinations
A character vector or list of character vectors proposing the assays or, in the case of a list, combination of assays to use with each element being a vector of assays to combine. Special value
"all"means all possible subsets of assays.- nFolds
A numeric specifying the number of folds to use for cross-validation.
- nRepeats
A numeric specifying the the number of repeats or permutations to use for cross-validation.
- nCores
A numeric specifying the number of cores used if the user wants to use parallelisation.
- characteristicsLabel
A character specifying an additional label for the cross-validation run.
- extraParams
A list of parameters that will be used to overwrite default settings of transformation, selection, or model-building functions or parameters which will be passed into the data cleaning function or cross-validation mode used for parameter tuning. Each name of a list element is a list and must be one of
"prepare","select","train","predict",tuneCross. By default, no parameter tuning is done. To use the a default parameter range for tuning (see the article titled Parameter Tuning Presets for crossValidate and Their Customisation on the website), specify a list element of"select"or"train"lists named"tuneParams"with value"auto". To specify your own range of values, specify alistwith names being the parameters in the functions described in the same article on the website. For the valid element names in the"prepare"list, see?prepareDatafor its parameter names. The list"tuneCross"can have elements named"tuneMode"and"performanceType". Valid values for"tuneMode"are"Resubstitution"or"Nested CV". For"performanceType", it is any of the metrics which can be specified tocalcPerformance.- verbose
Default: 0. A number between 0 and 3 for the amount of progress messages to give. A higher number will produce more messages as more lower-level functions print messages.
- x
Same as
measurementsbut only training samples.- outcomeTrain
For the
trainfunction, either a factor vector of classes, aSurvobject, or a character string, or vector of such strings, containing column name(s) of column(s) containing either classes or time and event information about survival. If column names of survival information, time must be in first column and event status in the second.- assayIDs
A character vector for assays to train with. Special value
"all"uses all assays in the input object.- object
A fitted model or a list of such models.
- newData
For the
predictfunction, an object of typematrix,data.frameDataFrame,list(of matrices or data frames) orMultiAssayExperimentcontaining the data to make predictions with with either a fitted model created bytrainor the final model stored in aClassifyResultobject.
Value
An object of class ClassifyResult
Details
classifier can be any a keyword for any of the implemented approaches as shown by available().
selectionMethod can be a keyword for any of the implemented approaches as shown by available("selectionMethod").
multiViewMethod can be a keyword for any of the implemented approaches as shown by available("multiViewMethod").
Examples
data(asthma)
# Compare randomForest and SVM classifiers.
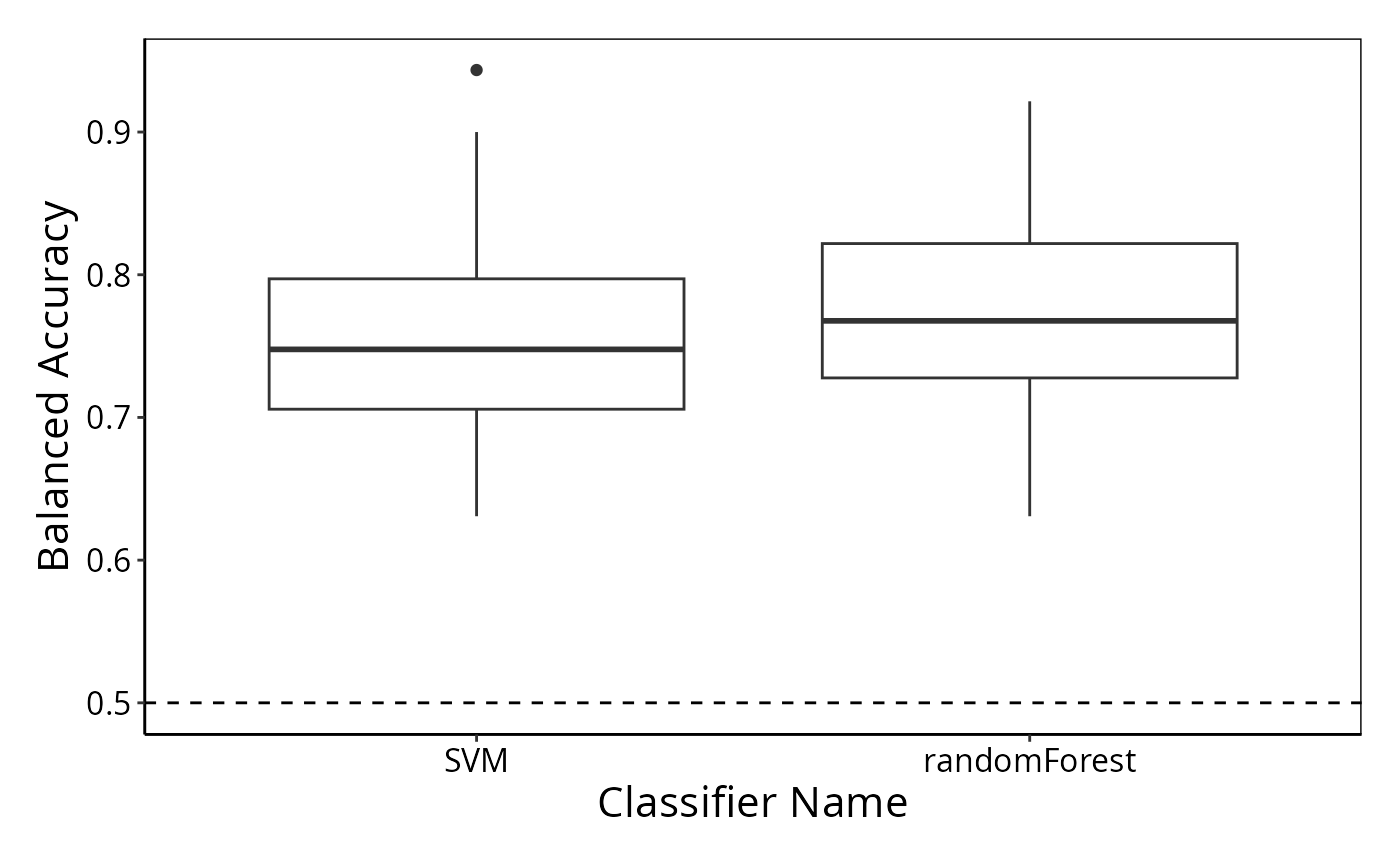
result <- crossValidate(measurements, classes, classifier = c("randomForest", "SVM"))
performancePlot(result)
#> Warning: Balanced Accuracy not found in all elements of results. Calculating it now.
 # Compare performance of different assays.
# First make a toy example assay with multiple data types. We'll randomly assign different features to be clinical, gene or protein.
# set.seed(51773)
# measurements <- DataFrame(measurements, check.names = FALSE)
# mcols(measurements)$assay <- c(rep("clinical", 20), sample(c("gene", "protein"), ncol(measurements) - 20, replace = TRUE))
# mcols(measurements)$feature <- colnames(measurements)
# We'll use different nFeatures for each assay. We'll also use repeated cross-validation with 5 repeats for speed in the example.
# set.seed(51773)
#result <- crossValidate(measurements, classes, nFeatures = c(clinical = 5, gene = 20, protein = 30), classifier = "randomForest", nRepeats = 5)
# performancePlot(result)
# Merge different assays. But we will only do this for two combinations. If assayCombinations is not specified it would attempt all combinations.
# set.seed(51773)
# resultMerge <- crossValidate(measurements, classes, assayCombinations = list(c("clinical", "protein"), c("clinical", "gene")), multiViewMethod = "merge", nRepeats = 5)
# performancePlot(resultMerge)
# performancePlot(c(result, resultMerge))
# Compare performance of different assays.
# First make a toy example assay with multiple data types. We'll randomly assign different features to be clinical, gene or protein.
# set.seed(51773)
# measurements <- DataFrame(measurements, check.names = FALSE)
# mcols(measurements)$assay <- c(rep("clinical", 20), sample(c("gene", "protein"), ncol(measurements) - 20, replace = TRUE))
# mcols(measurements)$feature <- colnames(measurements)
# We'll use different nFeatures for each assay. We'll also use repeated cross-validation with 5 repeats for speed in the example.
# set.seed(51773)
#result <- crossValidate(measurements, classes, nFeatures = c(clinical = 5, gene = 20, protein = 30), classifier = "randomForest", nRepeats = 5)
# performancePlot(result)
# Merge different assays. But we will only do this for two combinations. If assayCombinations is not specified it would attempt all combinations.
# set.seed(51773)
# resultMerge <- crossValidate(measurements, classes, assayCombinations = list(c("clinical", "protein"), c("clinical", "gene")), multiViewMethod = "merge", nRepeats = 5)
# performancePlot(resultMerge)
# performancePlot(c(result, resultMerge))