An introduction to CellSPA
Yingxin Lin
School of Mathematics and Statistics, The University of Sydney, AustraliaCellSPA.RmdRead BIDCell output
data_dir <- system.file("extdata/BIDCell_csv_output", package = "CellSPA")
data_dir
#> [1] "/dskh/nobackup/yingxinl/tmp/RtmprhTQ8f/temp_libpath22cabe42535ad0/CellSPA/extdata/BIDCell_csv_output"
tiff_path <- system.file("extdata/BIDCell_output_subset.tif", package = "CellSPA")
spe <- readBIDCell(data_dir,
tiff_path = tiff_path,
method_name = "BIDCell",
spatialCoordsNames = c("cell_centroid_x",
"cell_centroid_y"))
spe <- processingSPE(spe,
qc_range = list(total_transciprts = c(20, 2000),
total_genes = c(20, Inf)))
#subset a set of cells for illustration
spe <- CellSPA::subset(spe, 1:500)Baseline metrics
spe <- generatePolygon(spe)
#> | | | 0% | |======================================================================| 100%
spe <- calBaselineAllMetrics(spe, verbose = TRUE)
#> [1] "Metrics to run: total_transciprts, total_genes, total_cells, meanExprsPct_cells"
#> [1] "Calculating elongation"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating compactness"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating eccentricity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating sphericity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating solidity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating convexity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating circularity"
#> | | | 0% | |======================================================================| 100%
head(rowData(spe))
#> DataFrame with 6 rows and 2 columns
#> total_cells meanExprsPct_cells
#> <integer> <numeric>
#> SEC11C 248 0.496
#> DAPK3 54 0.108
#> TCIM 316 0.632
#> NKG7 16 0.032
#> RAPGEF3 69 0.138
#> PPARG 93 0.186
head(colData(spe))
#> DataFrame with 6 rows and 20 columns
#> cell_id pixel_size eccentricity cell_type spearman
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 27210 92 0.808957 7 0.578500
#> Cell_27211 27211 56 -Inf 6 0.408182
#> Cell_27212 27212 181 0.904051 7 0.569310
#> Cell_27213 27213 115 0.724503 7 0.474430
#> Cell_27214 27214 122 0.280000 7 0.538882
#> Cell_27215 27215 153 0.694516 7 0.546259
#> cell_type_atlas total_reads total_genes slide sample_id
#> <numeric> <numeric> <integer> <integer> <character>
#> Cell_27210 5 265 79 1 sample01
#> Cell_27211 41 74 40 1 sample01
#> Cell_27212 5 402 85 1 sample01
#> Cell_27213 42 289 92 1 sample01
#> Cell_27214 5 172 62 1 sample01
#> Cell_27215 42 259 67 1 sample01
#> total_transciprts sizeFactor cell_area elongation compactness
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 265 1.002193 92 0.727273 0.792508
#> Cell_27211 74 0.279858 56 0.433962 0.545574
#> Cell_27212 402 1.520309 181 0.928000 0.716283
#> Cell_27213 289 1.092958 115 0.923077 0.635395
#> Cell_27214 172 0.650480 122 0.336957 0.340099
#> Cell_27215 259 0.979502 153 0.839744 0.358405
#> sphericity solidity convexity circularity density
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.583612 0.949367 0.971781 0.839202 2.88043
#> Cell_27211 0.499965 0.847619 0.970686 0.579024 1.32143
#> Cell_27212 0.680052 0.883905 0.950348 0.793084 2.22099
#> Cell_27213 0.619852 0.840164 0.961928 0.686686 2.51304
#> Cell_27214 0.336094 0.731293 0.907927 0.412575 1.40984
#> Cell_27215 0.659214 0.615385 0.801868 0.557402 1.69281Expression similarity
Processing reference
sce_ref_full <- readRDS(system.file("extdata/sce_FFPE_full.rds",
package = "CellSPA"))
sce_ref <- processingRef(sce_ref_full,
celltype = sce_ref_full$graph_cluster_anno,
subset_row = rownames(spe))
sce_ref
#> class: SingleCellExperiment
#> dim: 307 17
#> metadata(0):
#> assays(2): mean prop_detected
#> rownames(307): SEC11C DAPK3 ... CD1C PDCD1
#> rowData names(3): ID Symbol Type
#> colnames(17): Macrophage Plasma ... LAMP3+ DC KRT15+ Myoepi
#> colData names(2): celltype Freq
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):Calculating expression similarity
spe <- calExpressionCorrelation(spe,
sce_ref,
ref_celltype = sce_ref$celltype,
method = c("pearson", "cosine"),
spe_exprs_values = "logcounts",
ref_exprs_values = "mean")
head(colData(spe))
#> DataFrame with 6 rows and 24 columns
#> cell_id pixel_size eccentricity cell_type spearman
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 27210 92 0.808957 7 0.578500
#> Cell_27211 27211 56 -Inf 6 0.408182
#> Cell_27212 27212 181 0.904051 7 0.569310
#> Cell_27213 27213 115 0.724503 7 0.474430
#> Cell_27214 27214 122 0.280000 7 0.538882
#> Cell_27215 27215 153 0.694516 7 0.546259
#> cell_type_atlas total_reads total_genes slide sample_id
#> <numeric> <numeric> <integer> <integer> <character>
#> Cell_27210 5 265 79 1 sample01
#> Cell_27211 41 74 40 1 sample01
#> Cell_27212 5 402 85 1 sample01
#> Cell_27213 42 289 92 1 sample01
#> Cell_27214 5 172 62 1 sample01
#> Cell_27215 42 259 67 1 sample01
#> total_transciprts sizeFactor cell_area elongation compactness
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 265 1.002193 92 0.727273 0.792508
#> Cell_27211 74 0.279858 56 0.433962 0.545574
#> Cell_27212 402 1.520309 181 0.928000 0.716283
#> Cell_27213 289 1.092958 115 0.923077 0.635395
#> Cell_27214 172 0.650480 122 0.336957 0.340099
#> Cell_27215 259 0.979502 153 0.839744 0.358405
#> sphericity solidity convexity circularity density
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.583612 0.949367 0.971781 0.839202 2.88043
#> Cell_27211 0.499965 0.847619 0.970686 0.579024 1.32143
#> Cell_27212 0.680052 0.883905 0.950348 0.793084 2.22099
#> Cell_27213 0.619852 0.840164 0.961928 0.686686 2.51304
#> Cell_27214 0.336094 0.731293 0.907927 0.412575 1.40984
#> Cell_27215 0.659214 0.615385 0.801868 0.557402 1.69281
#> mean_cor_correlation mean_celltype_correlation mean_cor_cosine
#> <numeric> <factor> <numeric>
#> Cell_27210 0.692426 CRABP2+ Malignant 0.753021
#> Cell_27211 0.609539 Firoblast 0.664884
#> Cell_27212 0.812256 CRABP2+ Malignant 0.849384
#> Cell_27213 0.652505 KRT15+ Myoepi 0.744028
#> Cell_27214 0.809740 CRABP2+ Malignant 0.842792
#> Cell_27215 0.789176 ECM1+ Malignant 0.826157
#> mean_celltype_cosine
#> <factor>
#> Cell_27210 CRABP2+ Malignant
#> Cell_27211 Firoblast
#> Cell_27212 CRABP2+ Malignant
#> Cell_27213 KRT15+ Myoepi
#> Cell_27214 CRABP2+ Malignant
#> Cell_27215 ECM1+ Malignant
spe <- calExpressionCorrelation(spe,
sce_ref,
ref_celltype = sce_ref$celltype,
method = c("pearson", "cosine"),
spe_exprs_values = "logcounts",
ref_exprs_values = "prop_detected")
head(colData(spe))
#> DataFrame with 6 rows and 28 columns
#> cell_id pixel_size eccentricity cell_type spearman
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 27210 92 0.808957 7 0.578500
#> Cell_27211 27211 56 -Inf 6 0.408182
#> Cell_27212 27212 181 0.904051 7 0.569310
#> Cell_27213 27213 115 0.724503 7 0.474430
#> Cell_27214 27214 122 0.280000 7 0.538882
#> Cell_27215 27215 153 0.694516 7 0.546259
#> cell_type_atlas total_reads total_genes slide sample_id
#> <numeric> <numeric> <integer> <integer> <character>
#> Cell_27210 5 265 79 1 sample01
#> Cell_27211 41 74 40 1 sample01
#> Cell_27212 5 402 85 1 sample01
#> Cell_27213 42 289 92 1 sample01
#> Cell_27214 5 172 62 1 sample01
#> Cell_27215 42 259 67 1 sample01
#> total_transciprts sizeFactor cell_area elongation compactness
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 265 1.002193 92 0.727273 0.792508
#> Cell_27211 74 0.279858 56 0.433962 0.545574
#> Cell_27212 402 1.520309 181 0.928000 0.716283
#> Cell_27213 289 1.092958 115 0.923077 0.635395
#> Cell_27214 172 0.650480 122 0.336957 0.340099
#> Cell_27215 259 0.979502 153 0.839744 0.358405
#> sphericity solidity convexity circularity density
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.583612 0.949367 0.971781 0.839202 2.88043
#> Cell_27211 0.499965 0.847619 0.970686 0.579024 1.32143
#> Cell_27212 0.680052 0.883905 0.950348 0.793084 2.22099
#> Cell_27213 0.619852 0.840164 0.961928 0.686686 2.51304
#> Cell_27214 0.336094 0.731293 0.907927 0.412575 1.40984
#> Cell_27215 0.659214 0.615385 0.801868 0.557402 1.69281
#> mean_cor_correlation mean_celltype_correlation mean_cor_cosine
#> <numeric> <factor> <numeric>
#> Cell_27210 0.692426 CRABP2+ Malignant 0.753021
#> Cell_27211 0.609539 Firoblast 0.664884
#> Cell_27212 0.812256 CRABP2+ Malignant 0.849384
#> Cell_27213 0.652505 KRT15+ Myoepi 0.744028
#> Cell_27214 0.809740 CRABP2+ Malignant 0.842792
#> Cell_27215 0.789176 ECM1+ Malignant 0.826157
#> mean_celltype_cosine prop_detected_cor_correlation
#> <factor> <numeric>
#> Cell_27210 CRABP2+ Malignant 0.683208
#> Cell_27211 Firoblast 0.571852
#> Cell_27212 CRABP2+ Malignant 0.786308
#> Cell_27213 KRT15+ Myoepi 0.628990
#> Cell_27214 CRABP2+ Malignant 0.777516
#> Cell_27215 ECM1+ Malignant 0.779704
#> prop_detected_celltype_correlation prop_detected_cor_cosine
#> <factor> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.756532
#> Cell_27211 Firoblast 0.625412
#> Cell_27212 SCGB2A2+ Malignant 0.828175
#> Cell_27213 KRT15+ Myoepi 0.734448
#> Cell_27214 CRABP2+ Malignant 0.806817
#> Cell_27215 ECM1+ Malignant 0.816152
#> prop_detected_celltype_cosine
#> <factor>
#> Cell_27210 SCGB2A2+ Malignant
#> Cell_27211 Firoblast
#> Cell_27212 SCGB2A2+ Malignant
#> Cell_27213 KRT15+ Myoepi
#> Cell_27214 CRABP2+ Malignant
#> Cell_27215 ECM1+ Malignant
spe <- calAggExpressionCorrelation(spe,
celltype = "mean_celltype_correlation",
sce_ref = sce_ref,
ref_celltype = "celltype",
method = c("pearson"),
spe_exprs_values = "logcounts",
ref_exprs_values = "mean")
diag(spe@metadata$CellSPA$similarity_metrics$agg_mean_correlation)
#> Macrophage Plasma VWF+ Endothelial ACTA2+ Myoepi
#> 0.7799986 0.7406420 0.7325303 0.7742887
#> CRABP2+ Malignant Firoblast CD4 T SCGB2A2+ Malignant
#> 0.7900731 0.7498582 0.7594411 0.7790271
#> ECM1+ Malignant CD163+ Macrophage STAB2+ Endothelial B Cells
#> 0.7261454 0.7260096 0.5062876 0.6522048
#> CD8 T IRF7+ DC Mast cells LAMP3+ DC
#> 0.7726590 0.0000000 0.5861591 0.5302542
#> KRT15+ Myoepi
#> 0.6805255
spe <- calAggExpressionCorrelation(spe,
celltype = "mean_celltype_correlation",
sce_ref = sce_ref,
ref_celltype = "celltype",
method = c("pearson"),
spe_exprs_values = "logcounts",
ref_exprs_values = "prop_detected")
diag(spe@metadata$CellSPA$similarity_metrics$agg_prop_detected_correlation)
#> Macrophage Plasma VWF+ Endothelial ACTA2+ Myoepi
#> 0.8344820 0.8234460 0.8169990 0.8803825
#> CRABP2+ Malignant Firoblast CD4 T SCGB2A2+ Malignant
#> 0.9192685 0.8389099 0.8055703 0.9168603
#> ECM1+ Malignant CD163+ Macrophage STAB2+ Endothelial B Cells
#> 0.9127056 0.8022071 0.5331824 0.6916012
#> CD8 T IRF7+ DC Mast cells LAMP3+ DC
#> 0.8034865 0.0000000 0.6200991 0.5597695
#> KRT15+ Myoepi
#> 0.7800218Calculate marker F1 purity
# Generate a positive marker list based on the reference data
positive_marker_list <- generateMarkerList(sce_ref, type = "positive")
#> [1] "For the top 10% of genes, the overlap freq between cell types"
#>
#> 1 2 3 4 5 6
#> 80 65 38 26 15 4
#> [1] "Length of the positive gene marker"
#> Macrophage Plasma VWF+ Endothelial ACTA2+ Myoepi
#> 27 28 30 21
#> CRABP2+ Malignant Firoblast CD4 T SCGB2A2+ Malignant
#> 21 27 26 22
#> ECM1+ Malignant CD163+ Macrophage STAB2+ Endothelial B Cells
#> 22 26 26 27
#> CD8 T IRF7+ DC Mast cells LAMP3+ DC
#> 27 24 27 25
#> KRT15+ Myoepi
#> 22
positive_marker_list
#> $Macrophage
#> [1] "ITGAX" "GLIPR1" "HAVCR2" "SCD" "PDK4" "CD68" "CD86" "S100A4"
#> [9] "C1QA" "AIF1" "MAP3K8" "CXCL16" "CD93" "LY86" "FCER1A" "MNDA"
#> [17] "FCER1G" "ITGAM" "SMAP2" "C1QC" "FGL2" "LYZ" "CD163" "APOC1"
#> [25] "FCGR3A" "CD14" "CD1C"
#>
#> $Plasma
#> [1] "SEC11C" "DUSP5" "SLAMF7" "ANKRD28" "MZB1" "CCPG1"
#> [7] "VOPP1" "PIM1" "ITM2C" "SEC24A" "SLAMF1" "PRDM1"
#> [13] "TPD52" "TUBA4A" "AQP3" "ERN1" "RAB30" "TENT5C"
#> [19] "CAV1" "DERL3" "TRIB1" "TNFRSF17" "CD79A" "CD19"
#> [25] "WARS" "CD79B" "TIFA" "CD27"
#>
#> $`VWF+ Endothelial`
#> [1] "TCIM" "RAPGEF3" "PPARG" "CLEC14A" "PDK4" "SOX18"
#> [7] "ACTA2" "PDGFRB" "CAVIN2" "RAMP2" "KDR" "EDNRB"
#> [13] "CD93" "EGFL7" "TCF4" "MMRN2" "POSTN" "CXCL12"
#> [19] "HOXD9" "VWF" "MMP2" "NDUFA4L2" "IL3RA" "ZEB1"
#> [25] "CLDN5" "ANKRD29" "CAV1" "AQP1" "MYLK" "NOSTRIN"
#>
#> $`ACTA2+ Myoepi`
#> [1] "S100A14" "CEACAM6" "KRT14" "ACTA2" "SFRP1" "ERBB2"
#> [7] "KRT5" "KRT6B" "TACSTD2" "PTN" "ACTG2" "SLC25A37"
#> [13] "DST" "SVIL" "EGFR" "OXTR" "CAV1" "SERPINA3"
#> [19] "DMKN" "MYLK" "MYH11"
#>
#> $`CRABP2+ Malignant`
#> [1] "TCIM" "S100A14" "CCDC6" "SCD" "CEACAM6" "CLDN4"
#> [7] "TRAF4" "TFAP2A" "MLPH" "ERBB2" "GATA3" "ANKRD30A"
#> [13] "EPCAM" "AGR3" "TACSTD2" "FOXA1" "LYPD3" "ELF3"
#> [19] "AR" "MYO5B" "SERPINA3"
#>
#> $Firoblast
#> [1] "TCIM" "LUM" "BASP1" "FBLN1" "ACTA2" "PDGFRB"
#> [7] "CRISPLD2" "EDNRB" "PDGFRA" "SFRP4" "DPT" "MEDAG"
#> [13] "TCF4" "ADH1B" "POSTN" "CXCL12" "PTGDS" "MMP2"
#> [19] "FSTL3" "SVIL" "ZEB1" "IGF1" "EGFR" "LRRC15"
#> [25] "PCOLCE" "AQP1" "CCDC80"
#>
#> $`CD4 T`
#> [1] "LTB" "TCF7" "CCL5" "S100A4" "LDHB" "GPR183" "TRAC"
#> [8] "CTLA4" "CCR7" "PTPRC" "SELL" "CD69" "SLAMF1" "CD247"
#> [15] "ADGRE5" "PRDM1" "FAM107B" "CD3G" "TUBA4A" "CD3E" "AQP3"
#> [22] "IL7R" "TIGIT" "CD3D" "KLRB1" "CD27"
#>
#> $`SCGB2A2+ Malignant`
#> [1] "S100A14" "SCD" "CEACAM6" "CLDN4" "TFAP2A" "MLPH"
#> [7] "ERBB2" "GATA3" "MZB1" "ANKRD30A" "EPCAM" "SDC4"
#> [13] "TACSTD2" "KLF5" "FOXA1" "TPD52" "ESR1" "LYPD3"
#> [19] "ELF3" "HOOK2" "AR" "SERPINA3"
#>
#> $`ECM1+ Malignant`
#> [1] "TCIM" "S100A14" "PCLAF" "CCDC6" "SCD" "CLDN4"
#> [7] "TRAF4" "USP53" "TFAP2A" "MLPH" "ERBB2" "GATA3"
#> [13] "MKI67" "ANKRD30A" "EPCAM" "FOXA1" "ELF3" "SH3YL1"
#> [19] "ABCC11" "AR" "MYO5B" "TOP2A"
#>
#> $`CD163+ Macrophage`
#> [1] "ITGAX" "GLIPR1" "HAVCR2" "PDK4" "CD68" "CD86" "C1QA" "AIF1"
#> [9] "MAP3K8" "MRC1" "CXCL16" "CD93" "ADAM9" "LY86" "FCER1G" "CXCL12"
#> [17] "ITGAM" "SMAP2" "IL2RA" "C1QC" "FGL2" "LYZ" "IGF1" "CD163"
#> [25] "FCGR3A" "CD14"
#>
#> $`STAB2+ Endothelial`
#> [1] "CLEC14A" "EDN1" "SOX18" "HOXD8" "CAVIN2" "RAMP2" "MRC1"
#> [8] "KDR" "FLNB" "EGFL7" "TCF4" "FAM107B" "HOXD9" "VWF"
#> [15] "MMP2" "DST" "SVIL" "ZEB1" "CLDN5" "IGF1" "CAV1"
#> [22] "BACE2" "FBLIM1" "LRRC15" "WARS" "CCDC80"
#>
#> $`B Cells`
#> [1] "GLIPR1" "CLECL1" "BASP1" "LTB" "GPR183" "CCR7" "PTPRC"
#> [8] "LY86" "SELL" "SLAMF1" "ADGRE5" "ITGAM" "FAM107B" "CD80"
#> [15] "MS4A1" "TPD52" "SMAP2" "TUBA4A" "SPIB" "BANK1" "RHOH"
#> [22] "CD79A" "CD19" "CD79B" "TIFA" "CD27" "CD1C"
#>
#> $`CD8 T`
#> [1] "NKG7" "KLRD1" "LTB" "GZMA" "TCF7" "CCL5" "DUSP2" "S100A4"
#> [9] "LAG3" "TRAC" "PTPRC" "PIM1" "ITM2C" "CD69" "GZMK" "CD247"
#> [17] "ADGRE5" "KLRC1" "PRF1" "CD3G" "TUBA4A" "CD3E" "CD8A" "IL7R"
#> [25] "TIGIT" "CD3D" "GNLY"
#>
#> $`IRF7+ DC`
#> [1] "GLIPR1" "DUSP5" "LTB" "CD68" "SLAMF7" "TRAF4" "GPR183" "MZB1"
#> [9] "PLD4" "KRT5" "LILRA4" "ITM2C" "SELL" "TCF4" "FCER1A" "FCER1G"
#> [17] "PTGDS" "SPIB" "TCL1A" "ERN1" "IL3RA" "DERL3" "TOMM7" "GZMB"
#>
#> $`Mast cells`
#> [1] "ITGAX" "HAVCR2" "PDE4A" "SERPINB9" "ANKRD28" "S100A4"
#> [7] "NPM3" "CAVIN2" "CD274" "MLPH" "HDC" "EDNRB"
#> [13] "CXCL16" "CD69" "FCER1A" "ADH1B" "ADGRE5" "FCER1G"
#> [19] "ITGAM" "FAM107B" "FSTL3" "CPA3" "RHOH" "BACE2"
#> [25] "KIT" "CTSG" "LIF"
#>
#> $`LAMP3+ DC`
#> [1] "FAM49A" "ITGAX" "CLECL1" "DUSP5" "BASP1" "CD83"
#> [7] "SERPINB9" "SLAMF7" "CD274" "MAP3K8" "GPR183" "CXCL16"
#> [13] "CCR7" "VOPP1" "CD80" "SMAP2" "SPIB" "PDCD1LG2"
#> [19] "LYZ" "ZEB1" "IL7R" "TRIB1" "WARS" "C15orf48"
#> [25] "PELI1"
#>
#> $`KRT15+ Myoepi`
#> [1] "KRT15" "KRT23" "DSC2" "CLDN4" "KRT14" "SFRP1"
#> [7] "TFAP2A" "KRT5" "EPCAM" "FLNB" "SDC4" "KRT6B"
#> [13] "TACSTD2" "PTN" "ELF3" "SLC25A37" "ALDH1A3" "DST"
#> [19] "SERPINA3" "ELF5" "PIGR" "SLC5A6"
negative_marker_list <- generateMarkerList(sce_ref, type = "negative", t = 1)
#> [1] "For the top 10% of genes, the overlap freq between cell types"
#>
#> 1 2 3 4 5 6 7 8 9 10 11 12 14
#> 22 9 4 4 4 8 3 6 7 15 3 4 2
#> [1] "Length of the positive gene marker"
#> Macrophage Plasma VWF+ Endothelial ACTA2+ Myoepi
#> 31 31 31 31
#> CRABP2+ Malignant Firoblast CD4 T SCGB2A2+ Malignant
#> 31 31 31 31
#> ECM1+ Malignant CD163+ Macrophage STAB2+ Endothelial B Cells
#> 31 31 31 31
#> CD8 T IRF7+ DC Mast cells LAMP3+ DC
#> 31 31 31 31
#> KRT15+ Myoepi
#> 31
negative_marker_list
#> $Macrophage
#> [1] "KRT15" "CEACAM6" "KRT14" "TRAF4" "ACTA2" "CAVIN2" "SFRP1"
#> [8] "CTTN" "HDC" "MZB1" "TRAC" "KRT5" "FLNB" "DSP"
#> [15] "CCR7" "EGFL7" "ITM2C" "TACSTD2" "MS4A1" "PTGDS" "VWF"
#> [22] "CPA3" "IL7R" "CAV1" "BACE2" "DERL3" "KIT" "CTSG"
#> [29] "AQP1" "MYLK" "GZMB"
#>
#> $Plasma
#> [1] "RUNX1" "ITGAX" "SCD" "PDK4" "CEACAM6" "LTB"
#> [7] "FASN" "CCND1" "TRAF4" "MLPH" "ERBB2" "CTTN"
#> [13] "GATA3" "ZEB2" "KRT5" "ANKRD30A" "DSP" "EGFL7"
#> [19] "TYROBP" "KRT7" "TACSTD2" "CD4" "ELF3" "CPA3"
#> [25] "CD9" "DST" "LYZ" "CD163" "IL7R" "KIT"
#> [31] "KRT8"
#>
#> $`VWF+ Endothelial`
#> [1] "CXCR4" "ITGAX" "SCD" "CEACAM6" "LTB" "FASN"
#> [7] "CLDN4" "MLPH" "ERBB2" "GATA3" "GPR183" "MZB1"
#> [13] "IL2RG" "KRT5" "ANKRD30A" "EPCAM" "DSP" "TYROBP"
#> [19] "KRT7" "ITM2C" "TACSTD2" "CD4" "CYTIP" "CDH1"
#> [25] "FOXA1" "ELF3" "CPA3" "CD9" "CD163" "IL7R"
#> [31] "KRT8"
#>
#> $`ACTA2+ Myoepi`
#> [1] "CXCR4" "ITGAX" "LTB" "CD68" "C1QA" "ZEB2" "HDC" "GPR183"
#> [9] "MZB1" "TRAC" "IL2RG" "EGFL7" "PTPRC" "TYROBP" "VOPP1" "ITM2C"
#> [17] "SELL" "TCF4" "ADGRE5" "FCER1G" "CD4" "CYTIP" "PTGDS" "SMAP2"
#> [25] "FGL2" "CPA3" "LYZ" "CD163" "IL7R" "KIT" "PECAM1"
#>
#> $`CRABP2+ Malignant`
#> [1] "CXCR4" "ITGAX" "GLIPR1" "PDK4" "LTB" "CD68" "S100A4" "C1QA"
#> [9] "ZEB2" "GPR183" "TRAC" "IL2RG" "PTPRC" "TYROBP" "ITM2C" "SELL"
#> [17] "TCF4" "ADGRE5" "FCER1G" "CD4" "CYTIP" "PTGDS" "SMAP2" "C1QC"
#> [25] "FGL2" "CPA3" "LYZ" "CD163" "IL7R" "KIT" "PECAM1"
#>
#> $Firoblast
#> [1] "SEC11C" "CXCR4" "ITGAX" "CEACAM6" "CD83" "LTB" "KRT14"
#> [8] "HDC" "GPR183" "MZB1" "IL2RG" "PLD4" "KRT5" "CCR7"
#> [15] "EGFL7" "PTPRC" "TYROBP" "SELL" "TACSTD2" "ADGRE5" "FCER1G"
#> [22] "CD4" "MS4A1" "CYTIP" "SMAP2" "CPA3" "CD9" "ERN1"
#> [29] "IL7R" "KIT" "PECAM1"
#>
#> $`CD4 T`
#> [1] "ITGAX" "S100A14" "SCD" "PDK4" "CEACAM6" "FASN"
#> [7] "CCND1" "CLDN4" "TRAF4" "MLPH" "ERBB2" "CTTN"
#> [13] "ZEB2" "MZB1" "KRT5" "ANKRD30A" "EPCAM" "DSP"
#> [19] "EGFL7" "KRT7" "ITM2C" "TACSTD2" "CDH1" "FOXA1"
#> [25] "ELF3" "CD9" "DST" "LYZ" "PECAM1" "KRT8"
#> [31] "ENAH"
#>
#> $`SCGB2A2+ Malignant`
#> [1] "CXCR4" "ITGAX" "GLIPR1" "PDK4" "LTB" "CD68" "C1QA" "ZEB2"
#> [9] "HDC" "GPR183" "TRAC" "IL2RG" "EGFL7" "TYROBP" "ITM2C" "SELL"
#> [17] "TCF4" "ADGRE5" "FCER1G" "CD4" "CYTIP" "PTGDS" "C1QC" "FGL2"
#> [25] "CPA3" "LYZ" "ERN1" "CD163" "IL7R" "KIT" "PECAM1"
#>
#> $`ECM1+ Malignant`
#> [1] "CXCR4" "ITGAX" "GLIPR1" "PDK4" "LTB" "CD68" "ACTA2" "C1QA"
#> [9] "ZEB2" "GPR183" "IL2RG" "KRT5" "TYROBP" "ITM2C" "SELL" "TCF4"
#> [17] "ADGRE5" "POSTN" "FCER1G" "CD4" "CYTIP" "PTGDS" "MMP2" "C1QC"
#> [25] "FGL2" "CPA3" "LYZ" "CD163" "IL7R" "KIT" "PECAM1"
#>
#> $`CD163+ Macrophage`
#> [1] "TCIM" "CXCR4" "S100A14" "SCD" "CEACAM6" "FASN"
#> [7] "CCND1" "CLDN4" "TRAF4" "ACTA2" "MLPH" "ERBB2"
#> [13] "CTTN" "GATA3" "MZB1" "KRT5" "ANKRD30A" "EPCAM"
#> [19] "FLNB" "DSP" "KRT7" "TACSTD2" "CDH1" "FOXA1"
#> [25] "ELF3" "CD9" "IL7R" "KIT" "TOMM7" "KRT8"
#> [31] "ENAH"
#>
#> $`STAB2+ Endothelial`
#> [1] "CXCR4" "RUNX1" "CEACAM6" "LTB" "FASN" "CLDN4"
#> [7] "TRAF4" "MLPH" "ERBB2" "GATA3" "ZEB2" "GPR183"
#> [13] "MZB1" "TRAC" "IL2RG" "KRT5" "ANKRD30A" "EPCAM"
#> [19] "TYROBP" "KRT7" "TACSTD2" "CD4" "CYTIP" "ELF3"
#> [25] "SMAP2" "CPA3" "LYZ" "CD163" "IL7R" "KIT"
#> [31] "KRT8"
#>
#> $`B Cells`
#> [1] "RUNX1" "PDK4" "CEACAM6" "FASN" "CCND1" "CLDN4"
#> [7] "ACTA2" "MLPH" "ERBB2" "CTTN" "GATA3" "KRT5"
#> [13] "ANKRD30A" "DSP" "EGFL7" "TYROBP" "KRT7" "ITM2C"
#> [19] "TACSTD2" "CD4" "FOXA1" "ELF3" "CPA3" "CD9"
#> [25] "DST" "LYZ" "CD163" "IL7R" "KIT" "PECAM1"
#> [31] "KRT8"
#>
#> $`CD8 T`
#> [1] "S100A14" "SCD" "PDK4" "CEACAM6" "FASN" "CCND1"
#> [7] "CLDN4" "TRAF4" "MLPH" "ERBB2" "CTTN" "MZB1"
#> [13] "KRT5" "ANKRD30A" "EPCAM" "DSP" "EGFL7" "KRT7"
#> [19] "TCF4" "TACSTD2" "CD4" "CDH1" "FOXA1" "ELF3"
#> [25] "CPA3" "CD9" "DST" "LYZ" "CD163" "KRT8"
#> [31] "ENAH"
#>
#> $`IRF7+ DC`
#> [1] "ITGAX" "SCD" "PDK4" "CEACAM6" "FASN" "CCND1"
#> [7] "CLDN4" "USP53" "ACTA2" "C1QA" "MLPH" "ERBB2"
#> [13] "CTTN" "GATA3" "HDC" "ANKRD30A" "DSP" "EGFL7"
#> [19] "KRT7" "TACSTD2" "ELF3" "C1QC" "FGL2" "CPA3"
#> [25] "CD9" "DST" "CD163" "IL7R" "CAV1" "KIT"
#> [31] "KRT8"
#>
#> $`Mast cells`
#> [1] "CXCR4" "S100A14" "SCD" "PDK4" "CEACAM6" "LTB"
#> [7] "FASN" "CCND1" "CLDN4" "TRAF4" "ACTA2" "ERBB2"
#> [13] "CTTN" "GATA3" "MZB1" "TRAC" "IL2RG" "ANKRD30A"
#> [19] "EPCAM" "DSP" "EGFL7" "KRT7" "TACSTD2" "CYTIP"
#> [25] "FOXA1" "ELF3" "DST" "IL7R" "TOMM7" "PECAM1"
#> [31] "KRT8"
#>
#> $`LAMP3+ DC`
#> [1] "TCIM" "SCD" "PDK4" "CEACAM6" "FASN" "CCND1"
#> [7] "CLDN4" "JUP" "TRAF4" "MLPH" "ERBB2" "CTTN"
#> [13] "GATA3" "MZB1" "KRT5" "ANKRD30A" "EPCAM" "DSP"
#> [19] "EGFL7" "KRT7" "ITM2C" "TACSTD2" "FCER1G" "FOXA1"
#> [25] "ELF3" "CPA3" "CD9" "CD163" "NARS" "KRT8"
#> [31] "CD14"
#>
#> $`KRT15+ Myoepi`
#> [1] "CXCR4" "ITGAX" "BASP1" "LTB" "CD68" "ZEB2" "HDC" "GPR183"
#> [9] "TRAC" "IL2RG" "CCR7" "EGFL7" "PTPRC" "TYROBP" "VOPP1" "ITM2C"
#> [17] "SELL" "TCF4" "CD69" "PRDM1" "POSTN" "CD4" "CYTIP" "PTGDS"
#> [25] "SMAP2" "FGL2" "CPA3" "LYZ" "CD163" "IL7R" "PECAM1"
spe <- calMarkerPurity(spe,
celltype = "mean_celltype_correlation",
marker_list = positive_marker_list,
marker_list_name = "positive")
head(colData(spe))
#> DataFrame with 6 rows and 31 columns
#> cell_id pixel_size eccentricity cell_type spearman
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 27210 92 0.808957 7 0.578500
#> Cell_27211 27211 56 -Inf 6 0.408182
#> Cell_27212 27212 181 0.904051 7 0.569310
#> Cell_27213 27213 115 0.724503 7 0.474430
#> Cell_27214 27214 122 0.280000 7 0.538882
#> Cell_27215 27215 153 0.694516 7 0.546259
#> cell_type_atlas total_reads total_genes slide sample_id
#> <numeric> <numeric> <integer> <integer> <character>
#> Cell_27210 5 265 79 1 sample01
#> Cell_27211 41 74 40 1 sample01
#> Cell_27212 5 402 85 1 sample01
#> Cell_27213 42 289 92 1 sample01
#> Cell_27214 5 172 62 1 sample01
#> Cell_27215 42 259 67 1 sample01
#> total_transciprts sizeFactor cell_area elongation compactness
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 265 1.002193 92 0.727273 0.792508
#> Cell_27211 74 0.279858 56 0.433962 0.545574
#> Cell_27212 402 1.520309 181 0.928000 0.716283
#> Cell_27213 289 1.092958 115 0.923077 0.635395
#> Cell_27214 172 0.650480 122 0.336957 0.340099
#> Cell_27215 259 0.979502 153 0.839744 0.358405
#> sphericity solidity convexity circularity density
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.583612 0.949367 0.971781 0.839202 2.88043
#> Cell_27211 0.499965 0.847619 0.970686 0.579024 1.32143
#> Cell_27212 0.680052 0.883905 0.950348 0.793084 2.22099
#> Cell_27213 0.619852 0.840164 0.961928 0.686686 2.51304
#> Cell_27214 0.336094 0.731293 0.907927 0.412575 1.40984
#> Cell_27215 0.659214 0.615385 0.801868 0.557402 1.69281
#> mean_cor_correlation mean_celltype_correlation mean_cor_cosine
#> <numeric> <factor> <numeric>
#> Cell_27210 0.692426 CRABP2+ Malignant 0.753021
#> Cell_27211 0.609539 Firoblast 0.664884
#> Cell_27212 0.812256 CRABP2+ Malignant 0.849384
#> Cell_27213 0.652505 KRT15+ Myoepi 0.744028
#> Cell_27214 0.809740 CRABP2+ Malignant 0.842792
#> Cell_27215 0.789176 ECM1+ Malignant 0.826157
#> mean_celltype_cosine prop_detected_cor_correlation
#> <factor> <numeric>
#> Cell_27210 CRABP2+ Malignant 0.683208
#> Cell_27211 Firoblast 0.571852
#> Cell_27212 CRABP2+ Malignant 0.786308
#> Cell_27213 KRT15+ Myoepi 0.628990
#> Cell_27214 CRABP2+ Malignant 0.777516
#> Cell_27215 ECM1+ Malignant 0.779704
#> prop_detected_celltype_correlation prop_detected_cor_cosine
#> <factor> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.756532
#> Cell_27211 Firoblast 0.625412
#> Cell_27212 SCGB2A2+ Malignant 0.828175
#> Cell_27213 KRT15+ Myoepi 0.734448
#> Cell_27214 CRABP2+ Malignant 0.806817
#> Cell_27215 ECM1+ Malignant 0.816152
#> prop_detected_celltype_cosine positive_F1 positive_Precision
#> <factor> <numeric> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.439024 0.450000
#> Cell_27211 Firoblast 0.324324 0.600000
#> Cell_27212 SCGB2A2+ Malignant 0.450000 0.473684
#> Cell_27213 KRT15+ Myoepi 0.210526 0.250000
#> Cell_27214 CRABP2+ Malignant 0.461538 0.500000
#> Cell_27215 ECM1+ Malignant 0.454545 0.454545
#> positive_Recall
#> <numeric>
#> Cell_27210 0.428571
#> Cell_27211 0.222222
#> Cell_27212 0.428571
#> Cell_27213 0.181818
#> Cell_27214 0.428571
#> Cell_27215 0.454545
spe <- calMarkerPurity(spe,
celltype = "mean_celltype_correlation",
marker_list = negative_marker_list,
marker_list_name = "negative")
head(colData(spe))
#> DataFrame with 6 rows and 34 columns
#> cell_id pixel_size eccentricity cell_type spearman
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 27210 92 0.808957 7 0.578500
#> Cell_27211 27211 56 -Inf 6 0.408182
#> Cell_27212 27212 181 0.904051 7 0.569310
#> Cell_27213 27213 115 0.724503 7 0.474430
#> Cell_27214 27214 122 0.280000 7 0.538882
#> Cell_27215 27215 153 0.694516 7 0.546259
#> cell_type_atlas total_reads total_genes slide sample_id
#> <numeric> <numeric> <integer> <integer> <character>
#> Cell_27210 5 265 79 1 sample01
#> Cell_27211 41 74 40 1 sample01
#> Cell_27212 5 402 85 1 sample01
#> Cell_27213 42 289 92 1 sample01
#> Cell_27214 5 172 62 1 sample01
#> Cell_27215 42 259 67 1 sample01
#> total_transciprts sizeFactor cell_area elongation compactness
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 265 1.002193 92 0.727273 0.792508
#> Cell_27211 74 0.279858 56 0.433962 0.545574
#> Cell_27212 402 1.520309 181 0.928000 0.716283
#> Cell_27213 289 1.092958 115 0.923077 0.635395
#> Cell_27214 172 0.650480 122 0.336957 0.340099
#> Cell_27215 259 0.979502 153 0.839744 0.358405
#> sphericity solidity convexity circularity density
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.583612 0.949367 0.971781 0.839202 2.88043
#> Cell_27211 0.499965 0.847619 0.970686 0.579024 1.32143
#> Cell_27212 0.680052 0.883905 0.950348 0.793084 2.22099
#> Cell_27213 0.619852 0.840164 0.961928 0.686686 2.51304
#> Cell_27214 0.336094 0.731293 0.907927 0.412575 1.40984
#> Cell_27215 0.659214 0.615385 0.801868 0.557402 1.69281
#> mean_cor_correlation mean_celltype_correlation mean_cor_cosine
#> <numeric> <factor> <numeric>
#> Cell_27210 0.692426 CRABP2+ Malignant 0.753021
#> Cell_27211 0.609539 Firoblast 0.664884
#> Cell_27212 0.812256 CRABP2+ Malignant 0.849384
#> Cell_27213 0.652505 KRT15+ Myoepi 0.744028
#> Cell_27214 0.809740 CRABP2+ Malignant 0.842792
#> Cell_27215 0.789176 ECM1+ Malignant 0.826157
#> mean_celltype_cosine prop_detected_cor_correlation
#> <factor> <numeric>
#> Cell_27210 CRABP2+ Malignant 0.683208
#> Cell_27211 Firoblast 0.571852
#> Cell_27212 CRABP2+ Malignant 0.786308
#> Cell_27213 KRT15+ Myoepi 0.628990
#> Cell_27214 CRABP2+ Malignant 0.777516
#> Cell_27215 ECM1+ Malignant 0.779704
#> prop_detected_celltype_correlation prop_detected_cor_cosine
#> <factor> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.756532
#> Cell_27211 Firoblast 0.625412
#> Cell_27212 SCGB2A2+ Malignant 0.828175
#> Cell_27213 KRT15+ Myoepi 0.734448
#> Cell_27214 CRABP2+ Malignant 0.806817
#> Cell_27215 ECM1+ Malignant 0.816152
#> prop_detected_celltype_cosine positive_F1 positive_Precision
#> <factor> <numeric> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.439024 0.450000
#> Cell_27211 Firoblast 0.324324 0.600000
#> Cell_27212 SCGB2A2+ Malignant 0.450000 0.473684
#> Cell_27213 KRT15+ Myoepi 0.210526 0.250000
#> Cell_27214 CRABP2+ Malignant 0.461538 0.500000
#> Cell_27215 ECM1+ Malignant 0.454545 0.454545
#> positive_Recall negative_F1 negative_Precision negative_Recall
#> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27211 0.222222 0.0000000 0.0000000 0.0000000
#> Cell_27212 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27213 0.181818 0.1333333 0.1379310 0.1290323
#> Cell_27214 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27215 0.454545 0.0333333 0.0344828 0.0322581Calculate marker expressed pct
spe <- calMarkerPct(spe,
celltype = "mean_celltype_correlation",
marker_list = positive_marker_list,
marker_list_name = "positive")
head(colData(spe))
#> DataFrame with 6 rows and 35 columns
#> cell_id pixel_size eccentricity cell_type spearman
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 27210 92 0.808957 7 0.578500
#> Cell_27211 27211 56 -Inf 6 0.408182
#> Cell_27212 27212 181 0.904051 7 0.569310
#> Cell_27213 27213 115 0.724503 7 0.474430
#> Cell_27214 27214 122 0.280000 7 0.538882
#> Cell_27215 27215 153 0.694516 7 0.546259
#> cell_type_atlas total_reads total_genes slide sample_id
#> <numeric> <numeric> <integer> <integer> <character>
#> Cell_27210 5 265 79 1 sample01
#> Cell_27211 41 74 40 1 sample01
#> Cell_27212 5 402 85 1 sample01
#> Cell_27213 42 289 92 1 sample01
#> Cell_27214 5 172 62 1 sample01
#> Cell_27215 42 259 67 1 sample01
#> total_transciprts sizeFactor cell_area elongation compactness
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 265 1.002193 92 0.727273 0.792508
#> Cell_27211 74 0.279858 56 0.433962 0.545574
#> Cell_27212 402 1.520309 181 0.928000 0.716283
#> Cell_27213 289 1.092958 115 0.923077 0.635395
#> Cell_27214 172 0.650480 122 0.336957 0.340099
#> Cell_27215 259 0.979502 153 0.839744 0.358405
#> sphericity solidity convexity circularity density
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.583612 0.949367 0.971781 0.839202 2.88043
#> Cell_27211 0.499965 0.847619 0.970686 0.579024 1.32143
#> Cell_27212 0.680052 0.883905 0.950348 0.793084 2.22099
#> Cell_27213 0.619852 0.840164 0.961928 0.686686 2.51304
#> Cell_27214 0.336094 0.731293 0.907927 0.412575 1.40984
#> Cell_27215 0.659214 0.615385 0.801868 0.557402 1.69281
#> mean_cor_correlation mean_celltype_correlation mean_cor_cosine
#> <numeric> <factor> <numeric>
#> Cell_27210 0.692426 CRABP2+ Malignant 0.753021
#> Cell_27211 0.609539 Firoblast 0.664884
#> Cell_27212 0.812256 CRABP2+ Malignant 0.849384
#> Cell_27213 0.652505 KRT15+ Myoepi 0.744028
#> Cell_27214 0.809740 CRABP2+ Malignant 0.842792
#> Cell_27215 0.789176 ECM1+ Malignant 0.826157
#> mean_celltype_cosine prop_detected_cor_correlation
#> <factor> <numeric>
#> Cell_27210 CRABP2+ Malignant 0.683208
#> Cell_27211 Firoblast 0.571852
#> Cell_27212 CRABP2+ Malignant 0.786308
#> Cell_27213 KRT15+ Myoepi 0.628990
#> Cell_27214 CRABP2+ Malignant 0.777516
#> Cell_27215 ECM1+ Malignant 0.779704
#> prop_detected_celltype_correlation prop_detected_cor_cosine
#> <factor> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.756532
#> Cell_27211 Firoblast 0.625412
#> Cell_27212 SCGB2A2+ Malignant 0.828175
#> Cell_27213 KRT15+ Myoepi 0.734448
#> Cell_27214 CRABP2+ Malignant 0.806817
#> Cell_27215 ECM1+ Malignant 0.816152
#> prop_detected_celltype_cosine positive_F1 positive_Precision
#> <factor> <numeric> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.439024 0.450000
#> Cell_27211 Firoblast 0.324324 0.600000
#> Cell_27212 SCGB2A2+ Malignant 0.450000 0.473684
#> Cell_27213 KRT15+ Myoepi 0.210526 0.250000
#> Cell_27214 CRABP2+ Malignant 0.461538 0.500000
#> Cell_27215 ECM1+ Malignant 0.454545 0.454545
#> positive_Recall negative_F1 negative_Precision negative_Recall
#> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27211 0.222222 0.0000000 0.0000000 0.0000000
#> Cell_27212 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27213 0.181818 0.1333333 0.1379310 0.1290323
#> Cell_27214 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27215 0.454545 0.0333333 0.0344828 0.0322581
#> positive_exprsPct
#> <numeric>
#> Cell_27210 0.809524
#> Cell_27211 0.444444
#> Cell_27212 0.904762
#> Cell_27213 0.681818
#> Cell_27214 1.000000
#> Cell_27215 0.772727
spe <- calMarkerPct(spe,
celltype = "mean_celltype_correlation",
marker_list = negative_marker_list,
marker_list_name = "negative")
head(colData(spe))
#> DataFrame with 6 rows and 36 columns
#> cell_id pixel_size eccentricity cell_type spearman
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 27210 92 0.808957 7 0.578500
#> Cell_27211 27211 56 -Inf 6 0.408182
#> Cell_27212 27212 181 0.904051 7 0.569310
#> Cell_27213 27213 115 0.724503 7 0.474430
#> Cell_27214 27214 122 0.280000 7 0.538882
#> Cell_27215 27215 153 0.694516 7 0.546259
#> cell_type_atlas total_reads total_genes slide sample_id
#> <numeric> <numeric> <integer> <integer> <character>
#> Cell_27210 5 265 79 1 sample01
#> Cell_27211 41 74 40 1 sample01
#> Cell_27212 5 402 85 1 sample01
#> Cell_27213 42 289 92 1 sample01
#> Cell_27214 5 172 62 1 sample01
#> Cell_27215 42 259 67 1 sample01
#> total_transciprts sizeFactor cell_area elongation compactness
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 265 1.002193 92 0.727273 0.792508
#> Cell_27211 74 0.279858 56 0.433962 0.545574
#> Cell_27212 402 1.520309 181 0.928000 0.716283
#> Cell_27213 289 1.092958 115 0.923077 0.635395
#> Cell_27214 172 0.650480 122 0.336957 0.340099
#> Cell_27215 259 0.979502 153 0.839744 0.358405
#> sphericity solidity convexity circularity density
#> <numeric> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.583612 0.949367 0.971781 0.839202 2.88043
#> Cell_27211 0.499965 0.847619 0.970686 0.579024 1.32143
#> Cell_27212 0.680052 0.883905 0.950348 0.793084 2.22099
#> Cell_27213 0.619852 0.840164 0.961928 0.686686 2.51304
#> Cell_27214 0.336094 0.731293 0.907927 0.412575 1.40984
#> Cell_27215 0.659214 0.615385 0.801868 0.557402 1.69281
#> mean_cor_correlation mean_celltype_correlation mean_cor_cosine
#> <numeric> <factor> <numeric>
#> Cell_27210 0.692426 CRABP2+ Malignant 0.753021
#> Cell_27211 0.609539 Firoblast 0.664884
#> Cell_27212 0.812256 CRABP2+ Malignant 0.849384
#> Cell_27213 0.652505 KRT15+ Myoepi 0.744028
#> Cell_27214 0.809740 CRABP2+ Malignant 0.842792
#> Cell_27215 0.789176 ECM1+ Malignant 0.826157
#> mean_celltype_cosine prop_detected_cor_correlation
#> <factor> <numeric>
#> Cell_27210 CRABP2+ Malignant 0.683208
#> Cell_27211 Firoblast 0.571852
#> Cell_27212 CRABP2+ Malignant 0.786308
#> Cell_27213 KRT15+ Myoepi 0.628990
#> Cell_27214 CRABP2+ Malignant 0.777516
#> Cell_27215 ECM1+ Malignant 0.779704
#> prop_detected_celltype_correlation prop_detected_cor_cosine
#> <factor> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.756532
#> Cell_27211 Firoblast 0.625412
#> Cell_27212 SCGB2A2+ Malignant 0.828175
#> Cell_27213 KRT15+ Myoepi 0.734448
#> Cell_27214 CRABP2+ Malignant 0.806817
#> Cell_27215 ECM1+ Malignant 0.816152
#> prop_detected_celltype_cosine positive_F1 positive_Precision
#> <factor> <numeric> <numeric>
#> Cell_27210 SCGB2A2+ Malignant 0.439024 0.450000
#> Cell_27211 Firoblast 0.324324 0.600000
#> Cell_27212 SCGB2A2+ Malignant 0.450000 0.473684
#> Cell_27213 KRT15+ Myoepi 0.210526 0.250000
#> Cell_27214 CRABP2+ Malignant 0.461538 0.500000
#> Cell_27215 ECM1+ Malignant 0.454545 0.454545
#> positive_Recall negative_F1 negative_Precision negative_Recall
#> <numeric> <numeric> <numeric> <numeric>
#> Cell_27210 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27211 0.222222 0.0000000 0.0000000 0.0000000
#> Cell_27212 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27213 0.181818 0.1333333 0.1379310 0.1290323
#> Cell_27214 0.428571 0.0000000 0.0000000 0.0000000
#> Cell_27215 0.454545 0.0333333 0.0344828 0.0322581
#> positive_exprsPct negative_exprsPct
#> <numeric> <numeric>
#> Cell_27210 0.809524 0.0967742
#> Cell_27211 0.444444 0.0967742
#> Cell_27212 0.904762 0.0322581
#> Cell_27213 0.681818 0.2580645
#> Cell_27214 1.000000 0.0322581
#> Cell_27215 0.772727 0.1612903Spatial Variation
spe <- calSpatialMetricsDiversity(spe,
celltype = "mean_celltype_correlation")
df <- spe@metadata$CellSPA$spatialMetricsDiversity$results
ggplot(df, aes(x = x_bin, y = y_bin, fill = `cellTypeProp_ECM1+ Malignant`)) +
geom_tile() +
scale_fill_viridis_c() +
theme(aspect.ratio = 1,
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank()) +
labs(fill = "ECM1+ Malignant %") 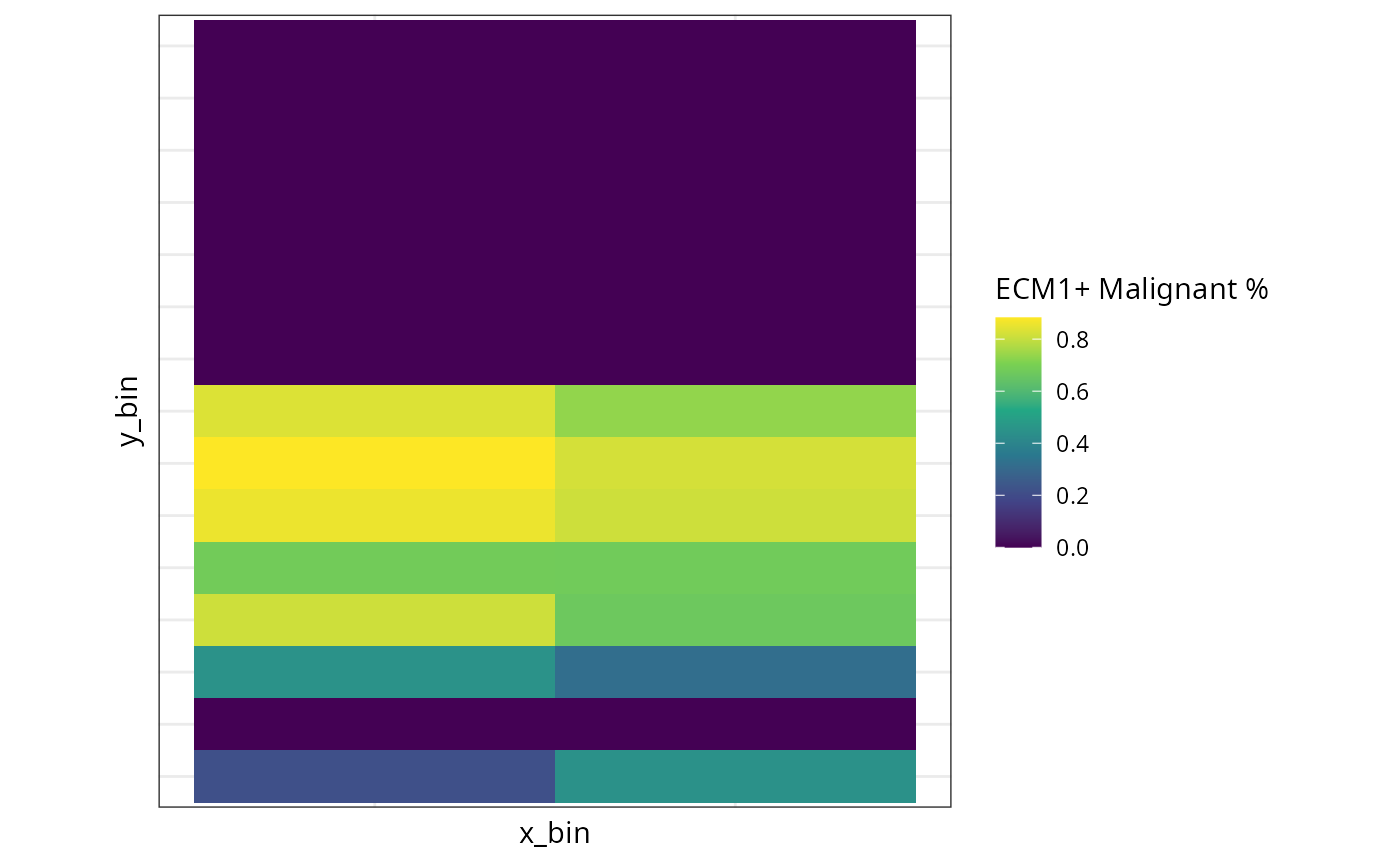
ggplot(df, aes(x = entropy, y = cv_total_transciprts,
color = `cellTypeProp_ECM1+ Malignant`)) +
geom_point() +
scale_color_viridis_c() +
theme(aspect.ratio = 1,
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank()) +
labs(color = "ECM1+ Malignant %")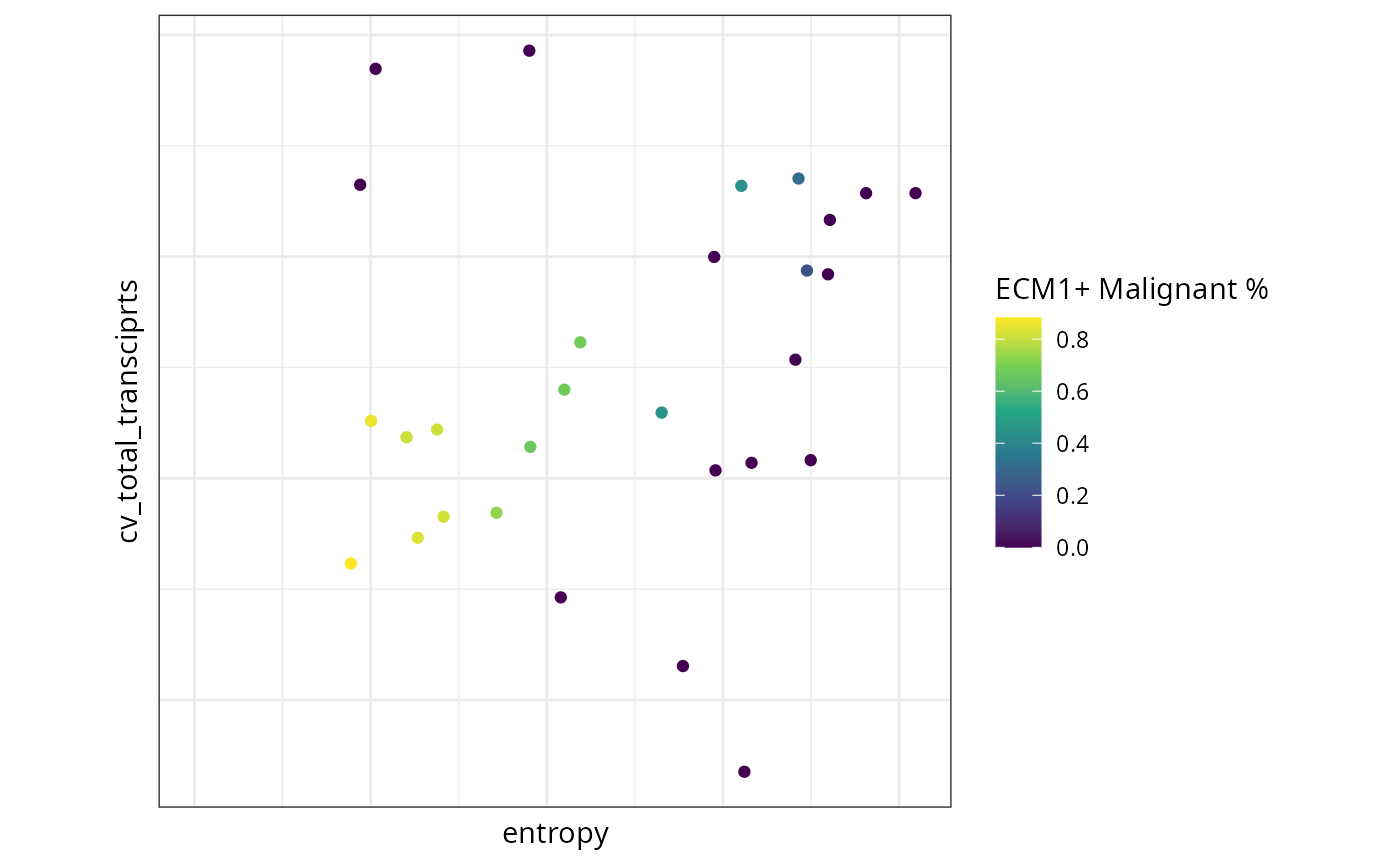
Neighbour purity
nn_celltype_pair <- c("B Cells", "CD4 T|CD8 T")
neg_markers <- list("B Cells" = c("CD3C", "CD3E", "CD8A"),
"CD4 T|CD8 T" = c("MS4A1", "CD79A", "CD79B"))
spe <- calNegMarkerVsDist(spe,
"mean_celltype_correlation",
nn_celltype_pair,
neg_markers)
spe@metadata$CellSPA$`negMarkerExprs_vs_dist`
#> $`B Cells`
#> CD3E CD8A
#> [0,10] NA NA
#> (10,20] 0.5 0
#> (20,30] NA NA
#> (30,40] 0.0 0
#> (40,50] 0.0 0
#> (50,100] 0.0 0
#> (100,334] 0.0 0
#>
#> $`CD4 T|CD8 T`
#> MS4A1 CD79A CD79B
#> [0,10] NA NA NA
#> (10,20] 0.00000000 0 0
#> (20,30] 0.00000000 0 0
#> (30,40] NA NA NA
#> (40,50] 0.00000000 0 0
#> (50,100] 0.00000000 0 0
#> (100,1.2e+03] 0.05263158 0 0Read 10x output
tenX_output_dir <- system.file("extdata/10x_output_subset", package = "CellSPA")
tenX_output_tif <- system.file("extdata/10x_output_subset/10x_from_csv_subset.tif", package = "CellSPA")
spe_10x <- readXenium(tenX_output_dir,
tiff_path = tenX_output_tif)
keep_idx <- spatialCoords(spe_10x)[, 2] <= max(spatialCoords(spe)[, 1]) &
spatialCoords(spe_10x)[, 2] >= min(spatialCoords(spe)[, 1])
spe_10x <- CellSPA::subset(spe_10x, which(keep_idx))
spe_10x <- processingSPE(spe_10x,
qc_range = list(total_transciprts = c(20, 2000),
total_genes = c(20, Inf)))
spe_10x <- CellSPA::subset(spe_10x, 1:500)
spe_10x
#> class: SpatialExperiment
#> dim: 313 500
#> metadata(2): CellSPA CellSegOutput
#> assays(2): counts logcounts
#> rownames(313): ABCC11 ACTA2 ... ZEB2 ZNF562
#> rowData names(5): ID Symbol Type total_cells meanExprsPct_cells
#> colnames(500): Cell_777 Cell_778 ... Cell_87976 Cell_88486
#> colData names(11): cell_id transcript_counts ... total_genes sizeFactor
#> reducedDimNames(2): PCA UMAP
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : x_centroid y_centroid
#> imgData names(1): sample_idRun CellSPA with a wrapper function CellSPA.
spe_10x <- CellSPA(spe_10x,
spe_celltype = NULL,
sce_ref = sce_ref,
ref_celltype = sce_ref$celltype,
positive_marker_list = positive_marker_list,
negative_marker_list = negative_marker_list,
nn_celltype_pair = nn_celltype_pair,
nn_neg_markers_list = neg_markers,
exprs_values = "logcounts",
use_BPPARAM = BiocParallel::SerialParam(),
verbose = TRUE)
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Metrics to run: total_transciprts, total_genes, total_cells, meanExprsPct_cells"
#> [1] "Calculating elongation"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating compactness"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating eccentricity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating sphericity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating solidity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating convexity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating circularity"
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating expression correlation metrics"
#> [1] "Calculating marker purity metrics"
#> | | | 0% | |======================================================================| 100%
#>
#> | | | 0% | |======================================================================| 100%
#>
#> | | | 0% | |======================================================================| 100%
#>
#> | | | 0% | |======================================================================| 100%
#>
#> [1] "Calculating spatial diversity metrics"
#> [1] "Calculating nn marker metrics"Visualisation
Within method
We can use plotColData in scater package to
visualise the baseline metrics.
scater::plotColData(spe, "total_transciprts",
x = "mean_celltype_correlation",
colour_by = "mean_celltype_correlation") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))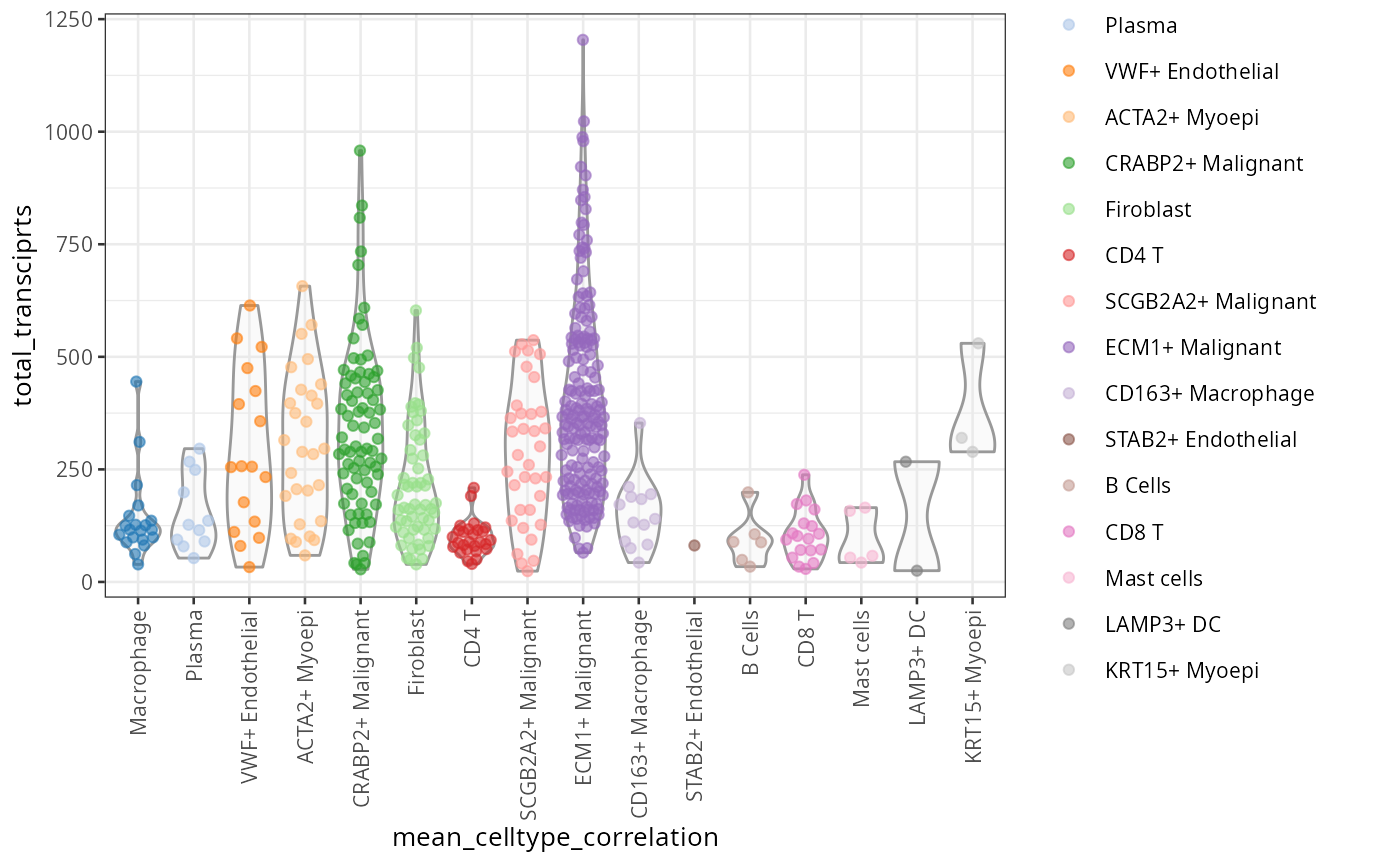
scater::plotColData(spe, "elongation",
x = "mean_celltype_correlation",
colour_by = "mean_celltype_correlation") +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5))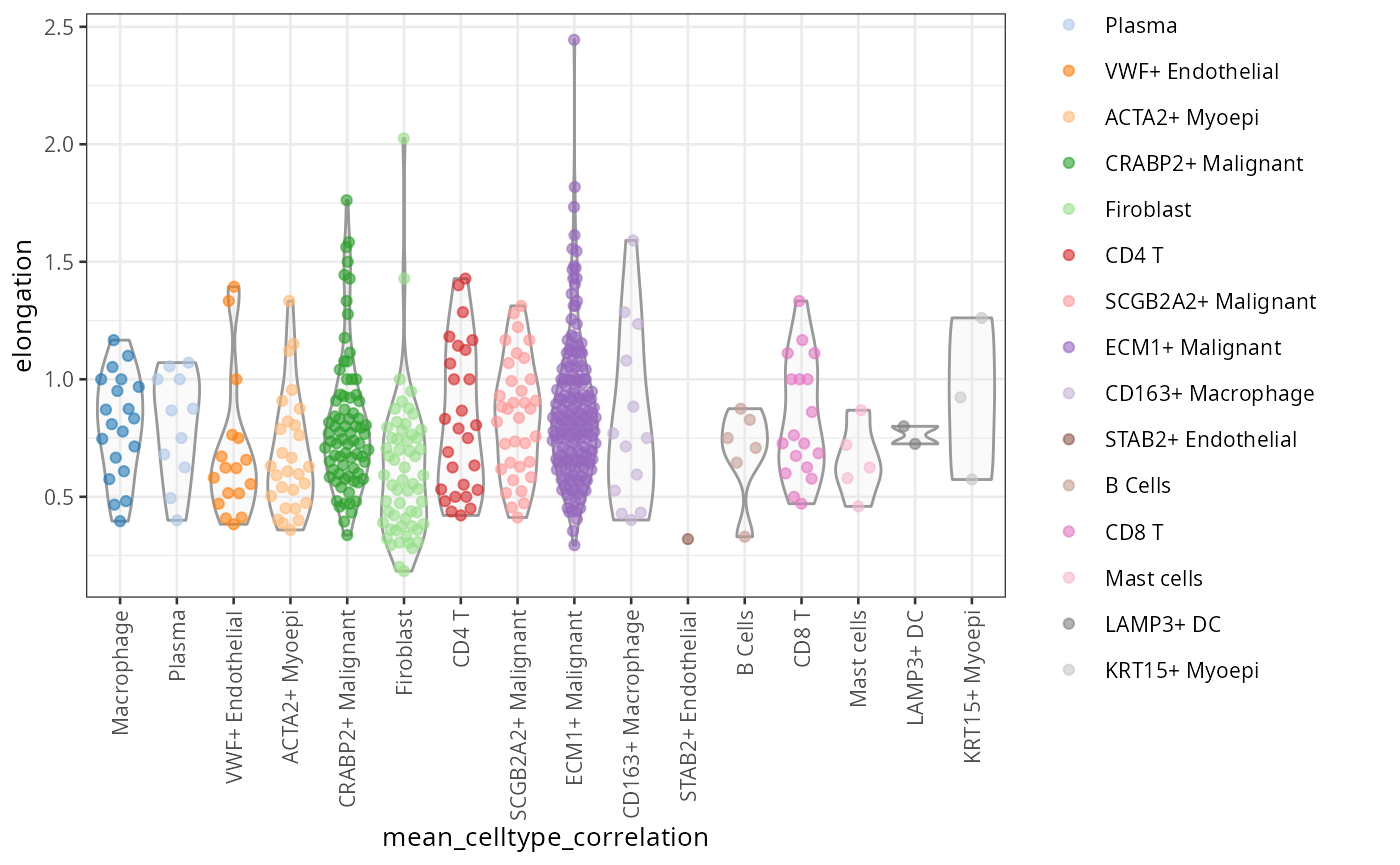
spe_list <- list(BIDCell = spe,
`10x` = spe_10x)Session Info
sessionInfo()
#> R version 4.4.1 (2024-06-14)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Debian GNU/Linux 12 (bookworm)
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.21.so; LAPACK version 3.11.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: Australia/Sydney
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] sf_1.0-17 SpatialExperiment_1.14.0
#> [3] scater_1.32.1 scuttle_1.14.0
#> [5] SingleCellExperiment_1.26.0 SummarizedExperiment_1.34.0
#> [7] Biobase_2.64.0 GenomicRanges_1.56.1
#> [9] GenomeInfoDb_1.40.1 IRanges_2.38.1
#> [11] S4Vectors_0.42.1 BiocGenerics_0.50.0
#> [13] MatrixGenerics_1.16.0 matrixStats_1.4.1
#> [15] ggthemes_5.1.0 ggplot2_3.5.1
#> [17] CellSPA_0.1.0 BiocStyle_2.32.1
#>
#> loaded via a namespace (and not attached):
#> [1] splines_4.4.1 tibble_3.2.1
#> [3] R.oo_1.26.0 polyclip_1.10-7
#> [5] lifecycle_1.0.4 edgeR_4.2.1
#> [7] lattice_0.22-6 MASS_7.3-61
#> [9] magrittr_2.0.3 limma_3.60.4
#> [11] sass_0.4.9 rmarkdown_2.28
#> [13] jquerylib_0.1.4 yaml_2.3.10
#> [15] sp_2.1-4 DBI_1.2.3
#> [17] multcomp_1.4-26 abind_1.4-5
#> [19] zlibbioc_1.50.0 purrr_1.0.2
#> [21] R.utils_2.12.3 Metrics_0.1.4
#> [23] TH.data_1.1-2 sandwich_3.1-0
#> [25] GenomeInfoDbData_1.2.12 ggrepel_0.9.6
#> [27] irlba_2.3.5.1 spatstat.utils_3.1-0
#> [29] terra_1.7-78 units_0.8-5
#> [31] spatstat.random_3.3-1 dqrng_0.4.1
#> [33] pkgdown_2.1.0 DelayedMatrixStats_1.26.0
#> [35] codetools_0.2-20 DropletUtils_1.24.0
#> [37] coin_1.4-3 DelayedArray_0.30.1
#> [39] alphahull_2.5 tidyselect_1.2.1
#> [41] raster_3.6-26 farver_2.1.2
#> [43] UCSC.utils_1.0.0 ScaledMatrix_1.12.0
#> [45] viridis_0.6.5 sgeostat_1.0-27
#> [47] jsonlite_1.8.8 BiocNeighbors_1.22.0
#> [49] e1071_1.7-16 survival_3.7-0
#> [51] systemfonts_1.1.0 tools_4.4.1
#> [53] ragg_1.3.3 Rcpp_1.0.13
#> [55] glue_1.7.0 shotGroups_0.8.2
#> [57] gridExtra_2.3 SparseArray_1.4.8
#> [59] xfun_0.47 dplyr_1.1.4
#> [61] HDF5Array_1.32.1 withr_3.0.1
#> [63] BiocManager_1.30.25 fastmap_1.2.0
#> [65] boot_1.3-31 rhdf5filters_1.16.0
#> [67] fansi_1.0.6 entropy_1.3.1
#> [69] digest_0.6.37 rsvd_1.0.5
#> [71] R6_2.5.1 textshaping_0.4.0
#> [73] colorspace_2.1-1 spatstat.data_3.1-2
#> [75] R.methodsS3_1.8.2 utf8_1.2.4
#> [77] generics_0.1.3 data.table_1.16.0
#> [79] FNN_1.1.4 class_7.3-22
#> [81] robustbase_0.99-4 httr_1.4.7
#> [83] htmlwidgets_1.6.4 S4Arrays_1.4.1
#> [85] uwot_0.2.2 pkgconfig_2.0.3
#> [87] gtable_0.3.5 modeltools_0.2-23
#> [89] XVector_0.44.0 htmltools_0.5.8.1
#> [91] bookdown_0.40 scales_1.3.0
#> [93] spatstat.univar_3.0-1 splancs_2.01-45
#> [95] knitr_1.48 rstudioapi_0.16.0
#> [97] reshape2_1.4.4 rjson_0.2.22
#> [99] proxy_0.4-27 cachem_1.1.0
#> [101] zoo_1.8-12 rhdf5_2.48.0
#> [103] stringr_1.5.1 KernSmooth_2.23-24
#> [105] parallel_4.4.1 vipor_0.4.7
#> [107] libcoin_1.0-10 desc_1.4.3
#> [109] pillar_1.9.0 grid_4.4.1
#> [111] proxyC_0.4.1 vctrs_0.6.5
#> [113] BiocSingular_1.20.0 beachmat_2.20.0
#> [115] beeswarm_0.4.0 evaluate_0.24.0
#> [117] magick_2.8.4 mvtnorm_1.3-1
#> [119] cli_3.6.3 locfit_1.5-9.10
#> [121] compiler_4.4.1 rlang_1.1.4
#> [123] crayon_1.5.3 labeling_0.4.3
#> [125] interp_1.1-6 classInt_0.4-10
#> [127] plyr_1.8.9 fs_1.6.4
#> [129] ggbeeswarm_0.7.2 stringi_1.8.4
#> [131] viridisLite_0.4.2 deldir_2.0-4
#> [133] BiocParallel_1.38.0 munsell_0.5.1
#> [135] tiff_0.1-12 spatstat.geom_3.3-2
#> [137] CompQuadForm_1.4.3 Matrix_1.7-0
#> [139] sparseMatrixStats_1.16.0 Rhdf5lib_1.26.0
#> [141] statmod_1.5.0 highr_0.11
#> [143] igraph_2.0.3 RcppParallel_5.1.9
#> [145] bslib_0.8.0 lwgeom_0.2-14
#> [147] DEoptimR_1.1-3